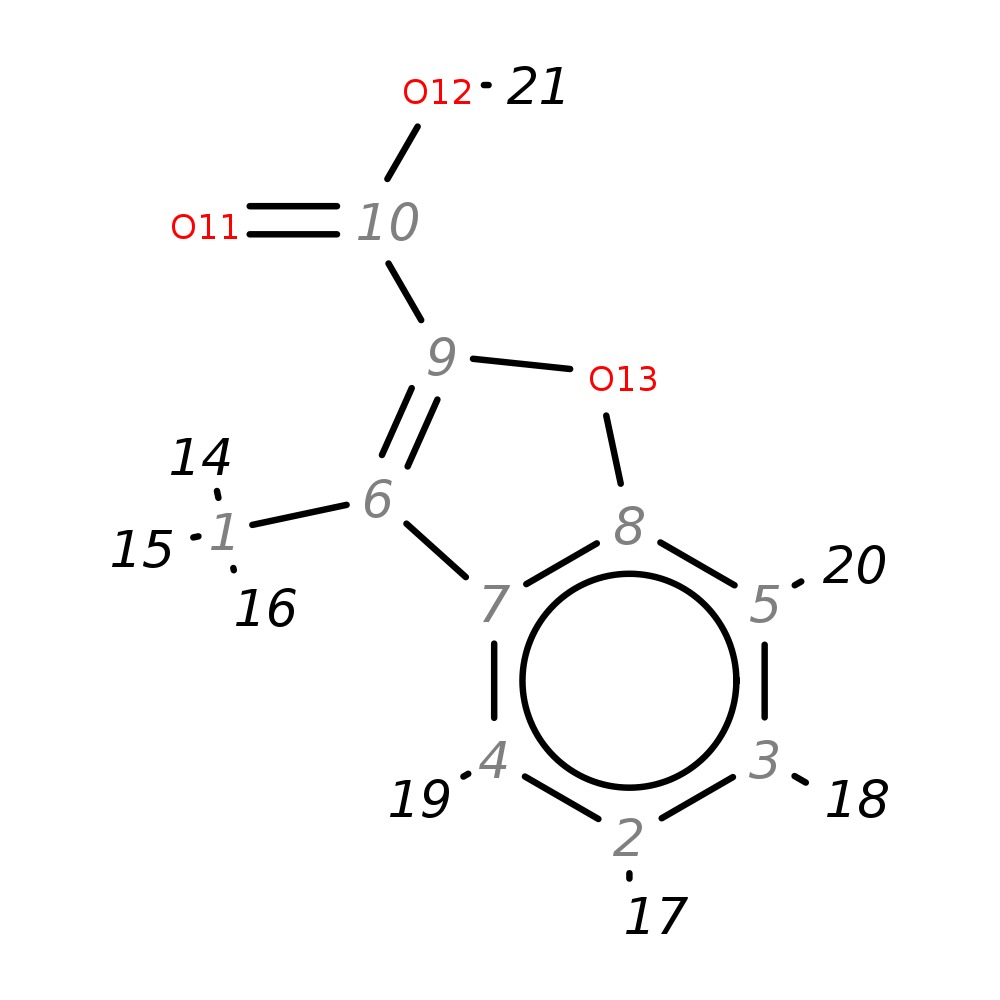
3-Methylbenzofuran-2-carboxylic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01813 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H8O3/c1-6-7-4-2-3-5-8(7)13-9(6)10(11)12/h2-5H,1H3,(H,11,12) | |
| Note 1 | 17,19?18,20 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Methylbenzofuran-2-carboxylic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|
| 14 | 2.524 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.524 | -14.0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.524 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.36 | 8.212 | 8.539 | 1.5 |
| 18 | 0 | 0 | 0 | 0 | 7.48 | 1.498 | 8.539 |
| 19 | 0 | 0 | 0 | 0 | 0 | 7.569 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.74 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.52389 | 1.0 | standard |
| 7.3444 | 0.0486922 | standard |
| 7.34623 | 0.0495261 | standard |
| 7.35823 | 0.109576 | standard |
| 7.36009 | 0.109793 | standard |
| 7.37209 | 0.0686637 | standard |
| 7.37409 | 0.0677641 | standard |
| 7.46514 | 0.0610003 | standard |
| 7.46696 | 0.0632188 | standard |
| 7.47913 | 0.110823 | standard |
| 7.48091 | 0.111783 | standard |
| 7.49293 | 0.056077 | standard |
| 7.49476 | 0.0552606 | standard |
| 7.56105 | 0.120773 | standard |
| 7.56304 | 0.118629 | standard |
| 7.57524 | 0.106653 | standard |
| 7.57715 | 0.105117 | standard |
| 7.7321 | 0.119408 | standard |
| 7.73393 | 0.119873 | standard |
| 7.74625 | 0.107261 | standard |
| 7.74816 | 0.106558 | standard |
| 2.52389 | 1.0 | standard |
| 7.05983 | 0.00796712 | standard |
| 7.1247 | 0.0134231 | standard |
| 7.25493 | 0.0426781 | standard |
| 7.37891 | 0.197701 | standard |
| 7.51033 | 0.444019 | standard |
| 7.60253 | 0.224723 | standard |
| 7.78379 | 0.0736331 | standard |
| 7.88655 | 0.0319551 | standard |
| 2.52389 | 1.0 | standard |
| 7.16804 | 0.0115071 | standard |
| 7.20305 | 0.0164372 | standard |
| 7.29282 | 0.0468272 | standard |
| 7.33461 | 0.068014 | standard |
| 7.3969 | 0.210452 | standard |
| 7.45233 | 0.200857 | standard |
| 7.5056 | 0.326837 | standard |
| 7.58376 | 0.128241 | standard |
| 7.60674 | 0.110115 | standard |
| 7.68133 | 0.164935 | standard |
| 7.79186 | 0.063201 | standard |
| 7.84062 | 0.0453812 | standard |
| 2.52389 | 1.0 | standard |
| 7.2212 | 0.0154521 | standard |
| 7.24431 | 0.0199462 | standard |
| 7.31841 | 0.0558743 | standard |
| 7.34506 | 0.0630411 | standard |
| 7.374 | 0.071461 | standard |
| 7.40703 | 0.18758 | standard |
| 7.43776 | 0.153461 | standard |
| 7.46649 | 0.0995142 | standard |
| 7.49899 | 0.198726 | standard |
| 7.52201 | 0.207426 | standard |
| 7.56264 | 0.12467 | standard |
| 7.60456 | 0.0802562 | standard |
| 7.64675 | 0.072908 | standard |
| 7.69095 | 0.134721 | standard |
| 7.71215 | 0.10901 | standard |
| 7.78182 | 0.0687753 | standard |
| 7.8129 | 0.057495 | standard |
| 2.52389 | 1.0 | standard |
| 7.23846 | 0.0170593 | standard |
| 7.2581 | 0.0216633 | standard |
| 7.32565 | 0.0608286 | standard |
| 7.34651 | 0.067933 | standard |
| 7.41283 | 0.167344 | standard |
| 7.43049 | 0.152382 | standard |
| 7.47128 | 0.0927801 | standard |
| 7.49632 | 0.168774 | standard |
| 7.52602 | 0.188076 | standard |
| 7.55719 | 0.133689 | standard |
| 7.5876 | 0.0611473 | standard |
| 7.60456 | 0.0713913 | standard |
| 7.63613 | 0.0707081 | standard |
| 7.69508 | 0.133735 | standard |
| 7.71447 | 0.113036 | standard |
| 7.77842 | 0.0711707 | standard |
| 7.80378 | 0.062411 | standard |
| 2.52389 | 1.0 | standard |
| 7.25205 | 0.0189651 | standard |
| 7.26891 | 0.0234033 | standard |
| 7.33092 | 0.0658894 | standard |
| 7.34861 | 0.073809 | standard |
| 7.40444 | 0.150596 | standard |
| 7.42324 | 0.178485 | standard |
| 7.47327 | 0.0912341 | standard |
| 7.4943 | 0.149994 | standard |
| 7.52923 | 0.177462 | standard |
| 7.55326 | 0.143858 | standard |
| 7.60322 | 0.070786 | standard |
| 7.62799 | 0.0703853 | standard |
| 7.69855 | 0.132845 | standard |
| 7.71607 | 0.115944 | standard |
| 7.77529 | 0.0742811 | standard |
| 7.79657 | 0.0673017 | standard |
| 2.52389 | 1.0 | standard |
| 7.30998 | 0.0327672 | standard |
| 7.31658 | 0.0347791 | standard |
| 7.35063 | 0.0964964 | standard |
| 7.35774 | 0.099593 | standard |
| 7.39138 | 0.0903928 | standard |
| 7.39885 | 0.0899842 | standard |
| 7.43624 | 0.0607371 | standard |
| 7.44439 | 0.0739786 | standard |
| 7.47787 | 0.10059 | standard |
| 7.48551 | 0.11468 | standard |
| 7.51897 | 0.0557024 | standard |
| 7.52669 | 0.0565457 | standard |
| 7.54654 | 0.139693 | standard |
| 7.55441 | 0.120581 | standard |
| 7.58779 | 0.0916733 | standard |
| 7.59556 | 0.0855442 | standard |
| 7.71733 | 0.126309 | standard |
| 7.72427 | 0.124087 | standard |
| 7.75896 | 0.094288 | standard |
| 7.76608 | 0.0915621 | standard |
| 2.52389 | 1.0 | standard |
| 7.32761 | 0.0396436 | standard |
| 7.33177 | 0.0409161 | standard |
| 7.35522 | 0.104656 | standard |
| 7.35936 | 0.105218 | standard |
| 7.38272 | 0.0796311 | standard |
| 7.38703 | 0.0783884 | standard |
| 7.45069 | 0.0607271 | standard |
| 7.45507 | 0.0678266 | standard |
| 7.47848 | 0.10498 | standard |
| 7.48292 | 0.111731 | standard |
| 7.50599 | 0.0528206 | standard |
| 7.51031 | 0.0516483 | standard |
| 7.55361 | 0.129328 | standard |
| 7.55803 | 0.120715 | standard |
| 7.58165 | 0.0985386 | standard |
| 7.58602 | 0.0945163 | standard |
| 7.72454 | 0.122312 | standard |
| 7.72869 | 0.122343 | standard |
| 7.75266 | 0.0997437 | standard |
| 7.75689 | 0.0988827 | standard |
| 2.52389 | 1.0 | standard |
| 7.3361 | 0.0437704 | standard |
| 7.3391 | 0.0448205 | standard |
| 7.35692 | 0.108041 | standard |
| 7.3598 | 0.108451 | standard |
| 7.37768 | 0.0744906 | standard |
| 7.38068 | 0.0733924 | standard |
| 7.45785 | 0.0602287 | standard |
| 7.46097 | 0.0648991 | standard |
| 7.4788 | 0.107773 | standard |
| 7.48185 | 0.111611 | standard |
| 7.49944 | 0.053579 | standard |
| 7.5026 | 0.0526135 | standard |
| 7.55727 | 0.124947 | standard |
| 7.5604 | 0.119642 | standard |
| 7.57846 | 0.102358 | standard |
| 7.58153 | 0.0997455 | standard |
| 7.72824 | 0.120426 | standard |
| 7.73124 | 0.120743 | standard |
| 7.74945 | 0.103503 | standard |
| 7.75244 | 0.102838 | standard |
| 2.52389 | 1.0 | standard |
| 7.34106 | 0.0463392 | standard |
| 7.34343 | 0.0472785 | standard |
| 7.35778 | 0.109481 | standard |
| 7.36004 | 0.109764 | standard |
| 7.37443 | 0.0711165 | standard |
| 7.37679 | 0.0701561 | standard |
| 7.46222 | 0.06048 | standard |
| 7.46457 | 0.0638535 | standard |
| 7.47897 | 0.109735 | standard |
| 7.4812 | 0.112142 | standard |
| 7.49553 | 0.0548621 | standard |
| 7.4979 | 0.0539838 | standard |
| 7.55951 | 0.123146 | standard |
| 7.56187 | 0.11966 | standard |
| 7.57646 | 0.104299 | standard |
| 7.5789 | 0.102358 | standard |
| 7.73054 | 0.119037 | standard |
| 7.73291 | 0.119477 | standard |
| 7.74748 | 0.105563 | standard |
| 7.74985 | 0.104896 | standard |
| 2.52389 | 1.0 | standard |
| 7.3444 | 0.0486922 | standard |
| 7.34623 | 0.0495261 | standard |
| 7.35823 | 0.109577 | standard |
| 7.36009 | 0.109794 | standard |
| 7.37209 | 0.0686647 | standard |
| 7.37409 | 0.0677641 | standard |
| 7.46514 | 0.0610013 | standard |
| 7.46696 | 0.0632197 | standard |
| 7.47913 | 0.110823 | standard |
| 7.48091 | 0.111784 | standard |
| 7.49293 | 0.056077 | standard |
| 7.49476 | 0.0552605 | standard |
| 7.56105 | 0.120774 | standard |
| 7.56304 | 0.11863 | standard |
| 7.57524 | 0.106653 | standard |
| 7.57715 | 0.105118 | standard |
| 7.7321 | 0.119409 | standard |
| 7.73393 | 0.119874 | standard |
| 7.74625 | 0.107262 | standard |
| 7.74816 | 0.106559 | standard |
| 2.52389 | 1.0 | standard |
| 7.34661 | 0.0494371 | standard |
| 7.34827 | 0.0502795 | standard |
| 7.35862 | 0.110248 | standard |
| 7.3601 | 0.11042 | standard |
| 7.37047 | 0.0673766 | standard |
| 7.37214 | 0.0665305 | standard |
| 7.46714 | 0.060054 | standard |
| 7.46879 | 0.0619024 | standard |
| 7.47918 | 0.110802 | standard |
| 7.48069 | 0.111636 | standard |
| 7.49098 | 0.0555717 | standard |
| 7.49264 | 0.0546666 | standard |
| 7.56211 | 0.119517 | standard |
| 7.56377 | 0.118447 | standard |
| 7.57431 | 0.107112 | standard |
| 7.57597 | 0.106006 | standard |
| 7.73312 | 0.116916 | standard |
| 7.73479 | 0.11746 | standard |
| 7.74531 | 0.108263 | standard |
| 7.74697 | 0.107625 | standard |
| 2.52389 | 1.0 | standard |
| 7.34747 | 0.0495188 | standard |
| 7.34909 | 0.0503774 | standard |
| 7.35875 | 0.111961 | standard |
| 7.36008 | 0.112104 | standard |
| 7.36975 | 0.0661995 | standard |
| 7.37138 | 0.0653408 | standard |
| 7.46802 | 0.0607314 | standard |
| 7.46948 | 0.0621267 | standard |
| 7.47915 | 0.110026 | standard |
| 7.48068 | 0.11073 | standard |
| 7.49029 | 0.0562332 | standard |
| 7.49175 | 0.0554064 | standard |
| 7.56262 | 0.119888 | standard |
| 7.56408 | 0.119426 | standard |
| 7.57388 | 0.106598 | standard |
| 7.57551 | 0.105505 | standard |
| 7.73362 | 0.11782 | standard |
| 7.73508 | 0.118341 | standard |
| 7.74501 | 0.109653 | standard |
| 7.74647 | 0.10906 | standard |
| 2.52389 | 1.0 | standard |
| 7.34837 | 0.0506698 | standard |
| 7.34978 | 0.0514778 | standard |
| 7.35883 | 0.111342 | standard |
| 7.3601 | 0.111481 | standard |
| 7.36916 | 0.0665197 | standard |
| 7.37058 | 0.0657118 | standard |
| 7.46869 | 0.0602187 | standard |
| 7.4701 | 0.0615763 | standard |
| 7.47927 | 0.111898 | standard |
| 7.48056 | 0.11229 | standard |
| 7.48957 | 0.0563093 | standard |
| 7.49098 | 0.0553577 | standard |
| 7.56299 | 0.119001 | standard |
| 7.5644 | 0.118797 | standard |
| 7.57359 | 0.108426 | standard |
| 7.575 | 0.107549 | standard |
| 7.73399 | 0.11697 | standard |
| 7.7354 | 0.117524 | standard |
| 7.74456 | 0.107341 | standard |
| 7.74613 | 0.10665 | standard |
| 2.52389 | 1.0 | standard |
| 7.34963 | 0.0514779 | standard |
| 7.3509 | 0.052258 | standard |
| 7.35903 | 0.112092 | standard |
| 7.36009 | 0.113219 | standard |
| 7.36828 | 0.0666768 | standard |
| 7.36938 | 0.0659861 | standard |
| 7.46994 | 0.0598983 | standard |
| 7.47121 | 0.0609392 | standard |
| 7.47925 | 0.112043 | standard |
| 7.48045 | 0.112235 | standard |
| 7.48845 | 0.0563671 | standard |
| 7.48972 | 0.0554723 | standard |
| 7.56366 | 0.117345 | standard |
| 7.56493 | 0.117655 | standard |
| 7.57307 | 0.109078 | standard |
| 7.57434 | 0.108255 | standard |
| 7.73466 | 0.116232 | standard |
| 7.73593 | 0.116817 | standard |
| 7.74407 | 0.109623 | standard |
| 7.74534 | 0.108985 | standard |
| 2.52389 | 1.0 | standard |
| 7.35021 | 0.0519568 | standard |
| 7.3514 | 0.0527309 | standard |
| 7.35902 | 0.111838 | standard |
| 7.36011 | 0.111951 | standard |
| 7.36774 | 0.0649973 | standard |
| 7.36893 | 0.0642361 | standard |
| 7.47043 | 0.0604066 | standard |
| 7.47161 | 0.0613931 | standard |
| 7.47935 | 0.111253 | standard |
| 7.48037 | 0.111298 | standard |
| 7.48805 | 0.0565678 | standard |
| 7.48924 | 0.0557145 | standard |
| 7.56385 | 0.117372 | standard |
| 7.56503 | 0.11767 | standard |
| 7.57287 | 0.109514 | standard |
| 7.57405 | 0.108744 | standard |
| 7.73495 | 0.115693 | standard |
| 7.73613 | 0.116705 | standard |
| 7.74386 | 0.110815 | standard |
| 7.74504 | 0.109752 | standard |
| 2.52389 | 1.0 | standard |
| 7.35071 | 0.0524894 | standard |
| 7.3518 | 0.0532221 | standard |
| 7.35912 | 0.112409 | standard |
| 7.36011 | 0.112514 | standard |
| 7.36744 | 0.0649517 | standard |
| 7.36853 | 0.0642227 | standard |
| 7.47092 | 0.0607937 | standard |
| 7.47201 | 0.0616127 | standard |
| 7.47935 | 0.11273 | standard |
| 7.48034 | 0.112739 | standard |
| 7.48765 | 0.0564764 | standard |
| 7.48874 | 0.055663 | standard |
| 7.56415 | 0.117648 | standard |
| 7.56523 | 0.118201 | standard |
| 7.57267 | 0.110277 | standard |
| 7.57375 | 0.109534 | standard |
| 7.73515 | 0.115903 | standard |
| 7.73623 | 0.11647 | standard |
| 7.74366 | 0.111715 | standard |
| 7.74475 | 0.111118 | standard |
| 2.52389 | 1.0 | standard |
| 7.35282 | 0.0533825 | standard |
| 7.35374 | 0.0541702 | standard |
| 7.35931 | 0.113476 | standard |
| 7.36003 | 0.112054 | standard |
| 7.3657 | 0.0621545 | standard |
| 7.36662 | 0.0613876 | standard |
| 7.47294 | 0.0596389 | standard |
| 7.47385 | 0.0604667 | standard |
| 7.47943 | 0.113566 | standard |
| 7.48017 | 0.111936 | standard |
| 7.48585 | 0.0576497 | standard |
| 7.48658 | 0.0569834 | standard |
| 7.56517 | 0.114156 | standard |
| 7.56609 | 0.114856 | standard |
| 7.57171 | 0.110372 | standard |
| 7.57263 | 0.109639 | standard |
| 7.73632 | 0.116936 | standard |
| 7.73705 | 0.117448 | standard |
| 7.74285 | 0.115111 | standard |
| 7.74358 | 0.114588 | standard |