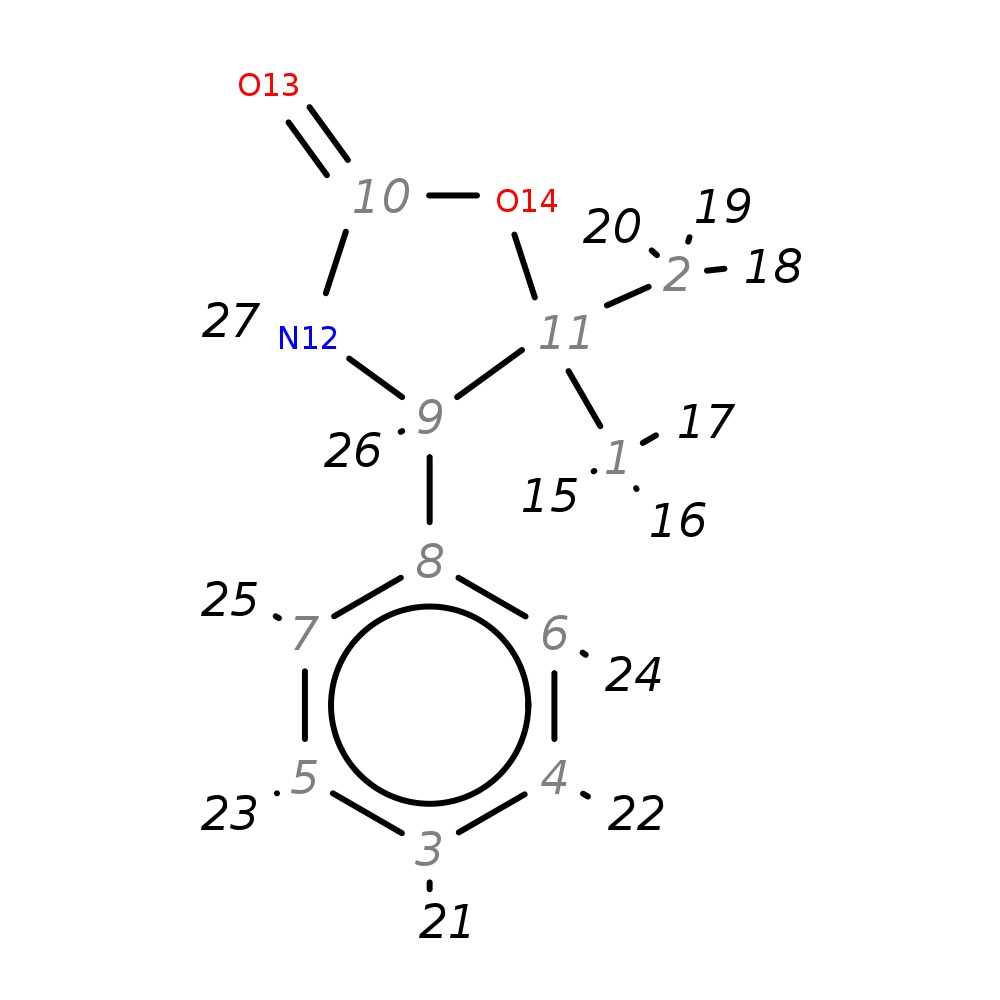
(R)-Phenyl superquat
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.18060 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H13NO2/c1-11(2)9(12-10(13)14-11)8-6-4-3-5-7-8/h3-7,9H,1-2H3,(H,12,13)/t9-/m0/s1 | |
| Note 1 | 26 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| (R)-Phenyl superquat | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 0.93 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0.93 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0.93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 1.631 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 1.631 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 1.631 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 7.426 | 9.067 | 9.067 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.464 | 1.5 | 7.86 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.464 | 0 | 7.86 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.363 | 1.5 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.363 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.37 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 0.930459 | 0.999544 | standard |
| 1.63121 | 1.0 | standard |
| 4.36984 | 0.334607 | standard |
| 7.356 | 0.231838 | standard |
| 7.3684 | 0.288302 | standard |
| 7.40818 | 0.0399464 | standard |
| 7.42299 | 0.0806521 | standard |
| 7.43714 | 0.157297 | standard |
| 7.45244 | 0.202627 | standard |
| 7.46446 | 0.211484 | standard |
| 7.4779 | 0.0612307 | standard |
| 0.930461 | 0.869819 | standard |
| 1.63121 | 0.869702 | standard |
| 4.36984 | 0.293353 | standard |
| 7.4172 | 1.0 | standard |
| 0.93046 | 1.0 | standard |
| 1.63121 | 0.998931 | standard |
| 4.36984 | 0.336221 | standard |
| 7.26227 | 0.0423511 | standard |
| 7.42361 | 0.868261 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.998937 | standard |
| 4.36984 | 0.33534 | standard |
| 7.295 | 0.0554933 | standard |
| 7.39708 | 0.505694 | standard |
| 7.42668 | 0.675608 | standard |
| 7.52055 | 0.0543832 | standard |
| 7.63818 | 0.00731113 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.99986 | standard |
| 4.36984 | 0.335043 | standard |
| 7.30571 | 0.0628251 | standard |
| 7.39429 | 0.450058 | standard |
| 7.42857 | 0.594665 | standard |
| 7.50894 | 0.0611913 | standard |
| 7.61805 | 0.00758012 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.999475 | standard |
| 4.36984 | 0.334458 | standard |
| 7.31513 | 0.0690642 | standard |
| 7.35487 | 0.154292 | standard |
| 7.39299 | 0.402552 | standard |
| 7.43136 | 0.533118 | standard |
| 7.59881 | 0.00954913 | standard |
| 0.930459 | 0.999459 | standard |
| 1.63121 | 1.0 | standard |
| 4.36984 | 0.334083 | standard |
| 7.34251 | 0.126817 | standard |
| 7.35386 | 0.100645 | standard |
| 7.37829 | 0.24043 | standard |
| 7.4107 | 0.086437 | standard |
| 7.44254 | 0.436265 | standard |
| 7.47208 | 0.156211 | standard |
| 7.50065 | 0.038611 | standard |
| 7.52565 | 0.018812 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.999991 | standard |
| 4.36984 | 0.334302 | standard |
| 7.34923 | 0.169304 | standard |
| 7.37369 | 0.242474 | standard |
| 7.39967 | 0.0229131 | standard |
| 7.41815 | 0.0617071 | standard |
| 7.42578 | 0.07555 | standard |
| 7.44513 | 0.350852 | standard |
| 7.46741 | 0.17666 | standard |
| 7.48965 | 0.042476 | standard |
| 7.5025 | 0.0293431 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.996573 | standard |
| 4.36984 | 0.333989 | standard |
| 7.35285 | 0.199272 | standard |
| 7.37101 | 0.262791 | standard |
| 7.39864 | 0.0337274 | standard |
| 7.42056 | 0.0631058 | standard |
| 7.42536 | 0.0654029 | standard |
| 7.44053 | 0.210294 | standard |
| 7.44876 | 0.242472 | standard |
| 7.46554 | 0.186205 | standard |
| 7.48393 | 0.0479485 | standard |
| 7.49132 | 0.0384053 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.997378 | standard |
| 4.36984 | 0.334307 | standard |
| 7.35476 | 0.219456 | standard |
| 7.36943 | 0.281971 | standard |
| 7.40438 | 0.0364567 | standard |
| 7.422 | 0.0712823 | standard |
| 7.42517 | 0.0665832 | standard |
| 7.43859 | 0.170662 | standard |
| 7.45085 | 0.217893 | standard |
| 7.46467 | 0.200894 | standard |
| 7.48027 | 0.0542809 | standard |
| 7.48461 | 0.0473122 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.998103 | standard |
| 4.36984 | 0.334366 | standard |
| 7.356 | 0.231672 | standard |
| 7.3684 | 0.288096 | standard |
| 7.40818 | 0.0399174 | standard |
| 7.42299 | 0.0805941 | standard |
| 7.43714 | 0.157186 | standard |
| 7.45244 | 0.202483 | standard |
| 7.46446 | 0.211333 | standard |
| 7.4779 | 0.0611869 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.999879 | standard |
| 4.36984 | 0.334223 | standard |
| 7.35693 | 0.237603 | standard |
| 7.36765 | 0.293047 | standard |
| 7.41085 | 0.0439 | standard |
| 7.42361 | 0.0895727 | standard |
| 7.43596 | 0.148791 | standard |
| 7.45377 | 0.196254 | standard |
| 7.46443 | 0.217517 | standard |
| 7.47626 | 0.067578 | standard |
| 0.930459 | 0.99815 | standard |
| 1.63121 | 1.0 | standard |
| 4.36984 | 0.333917 | standard |
| 7.35736 | 0.242386 | standard |
| 7.36735 | 0.291621 | standard |
| 7.41191 | 0.0458842 | standard |
| 7.42385 | 0.0945572 | standard |
| 7.43545 | 0.144154 | standard |
| 7.45429 | 0.192151 | standard |
| 7.46437 | 0.216764 | standard |
| 7.47573 | 0.0720552 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.998333 | standard |
| 4.36984 | 0.333769 | standard |
| 7.35764 | 0.24331 | standard |
| 7.3671 | 0.291602 | standard |
| 7.41282 | 0.0477337 | standard |
| 7.42403 | 0.0982258 | standard |
| 7.43496 | 0.141106 | standard |
| 7.45482 | 0.190199 | standard |
| 7.46436 | 0.220234 | standard |
| 7.47514 | 0.0752183 | standard |
| 0.930459 | 0.999924 | standard |
| 1.63121 | 1.0 | standard |
| 4.36984 | 0.333188 | standard |
| 7.35821 | 0.248995 | standard |
| 7.36668 | 0.292391 | standard |
| 7.41436 | 0.0512176 | standard |
| 7.42434 | 0.105436 | standard |
| 7.4341 | 0.135284 | standard |
| 7.45568 | 0.183032 | standard |
| 7.46426 | 0.219166 | standard |
| 7.47398 | 0.080938 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.999122 | standard |
| 4.36984 | 0.333056 | standard |
| 7.35845 | 0.250377 | standard |
| 7.36648 | 0.292176 | standard |
| 7.41497 | 0.0527281 | standard |
| 7.42447 | 0.108421 | standard |
| 7.43373 | 0.132931 | standard |
| 7.45607 | 0.181808 | standard |
| 7.46424 | 0.221481 | standard |
| 7.47341 | 0.086768 | standard |
| 0.930459 | 0.999306 | standard |
| 1.63121 | 1.0 | standard |
| 4.36984 | 0.332707 | standard |
| 7.35865 | 0.252522 | standard |
| 7.36629 | 0.292203 | standard |
| 7.41557 | 0.0538961 | standard |
| 7.42457 | 0.110969 | standard |
| 7.43341 | 0.131466 | standard |
| 7.4564 | 0.176737 | standard |
| 7.46421 | 0.21892 | standard |
| 7.47289 | 0.0866392 | standard |
| 0.930459 | 1.0 | standard |
| 1.63121 | 0.999258 | standard |
| 4.36984 | 0.332832 | standard |
| 7.35962 | 0.260538 | standard |
| 7.36548 | 0.292939 | standard |
| 7.41799 | 0.0600816 | standard |
| 7.4249 | 0.127458 | standard |
| 7.43182 | 0.119081 | standard |
| 7.45803 | 0.173634 | standard |
| 7.46411 | 0.220268 | standard |
| 7.4708 | 0.101623 | standard |