4'-Methoxyacetanilide
Simulation outputs:
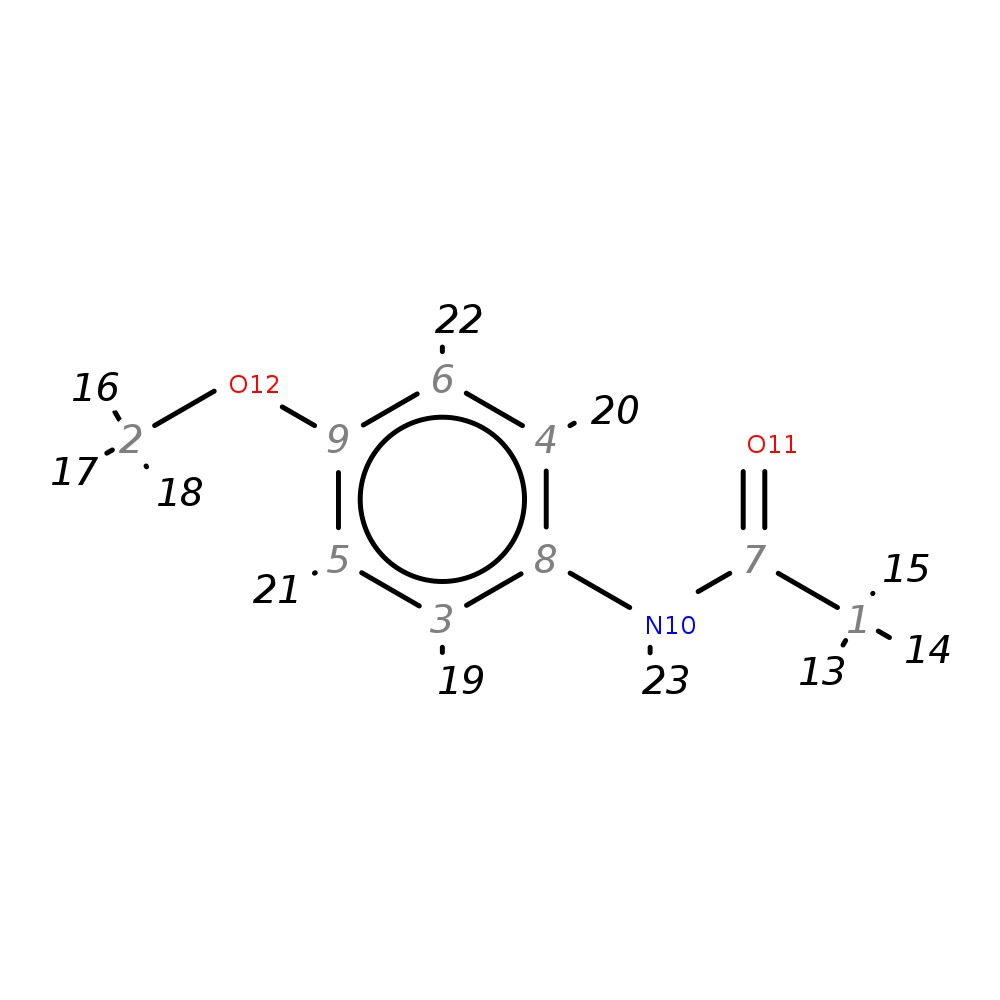
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01697 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H11NO2/c1-7(11)10-8-3-5-9(12-2)6-4-8/h3-6H,1-2H3,(H,10,11) | |
| Note 1 | 13,14,15?16,17,18 | |
| Note 2 | 19,20?21,22 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4'-Methoxyacetanilide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 2.143 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.143 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.143 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 3.83 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 3.83 | -14.0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 3.83 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 7.325 | 1.5 | 9.193 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.325 | 0 | 9.193 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.009 | 1.5 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.009 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.999899 | standard |
| 7.00112 | 0.269324 | standard |
| 7.01638 | 0.296162 | standard |
| 7.31772 | 0.29621 | standard |
| 7.33288 | 0.269055 | standard |
| 2.14252 | 0.999652 | standard |
| 3.82978 | 1.0 | standard |
| 6.85497 | 0.121262 | standard |
| 7.08861 | 0.467335 | standard |
| 7.24538 | 0.46737 | standard |
| 7.47904 | 0.121195 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.998566 | standard |
| 6.91442 | 0.163132 | standard |
| 7.06848 | 0.407605 | standard |
| 7.26554 | 0.407696 | standard |
| 7.41969 | 0.163078 | standard |
| 2.14252 | 0.998968 | standard |
| 3.82978 | 1.0 | standard |
| 6.94138 | 0.188711 | standard |
| 7.0564 | 0.378396 | standard |
| 7.27767 | 0.378459 | standard |
| 7.39272 | 0.188589 | standard |
| 2.14252 | 0.998865 | standard |
| 3.82978 | 1.0 | standard |
| 6.94989 | 0.198808 | standard |
| 7.05195 | 0.36735 | standard |
| 7.28204 | 0.367237 | standard |
| 7.38413 | 0.198816 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.998901 | standard |
| 6.95657 | 0.206496 | standard |
| 7.04832 | 0.359153 | standard |
| 7.28578 | 0.359152 | standard |
| 7.37752 | 0.206494 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.999109 | standard |
| 6.98444 | 0.243302 | standard |
| 7.03011 | 0.32176 | standard |
| 7.30392 | 0.321418 | standard |
| 7.34966 | 0.243291 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.999851 | standard |
| 6.99293 | 0.256192 | standard |
| 7.02341 | 0.309333 | standard |
| 7.31062 | 0.308873 | standard |
| 7.34106 | 0.256616 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.999915 | standard |
| 6.99707 | 0.262379 | standard |
| 7.01993 | 0.302273 | standard |
| 7.31418 | 0.303214 | standard |
| 7.33692 | 0.262987 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.9982 | standard |
| 6.99954 | 0.266613 | standard |
| 7.01776 | 0.298785 | standard |
| 7.31624 | 0.297642 | standard |
| 7.33456 | 0.265587 | standard |
| 2.14252 | 0.998805 | standard |
| 3.82978 | 1.0 | standard |
| 7.00112 | 0.269177 | standard |
| 7.01638 | 0.296001 | standard |
| 7.31772 | 0.296049 | standard |
| 7.33288 | 0.268909 | standard |
| 2.14252 | 0.999497 | standard |
| 3.82978 | 1.0 | standard |
| 7.0023 | 0.27119 | standard |
| 7.0153 | 0.294186 | standard |
| 7.3187 | 0.292631 | standard |
| 7.3318 | 0.271141 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.99972 | standard |
| 7.00269 | 0.271772 | standard |
| 7.0149 | 0.293236 | standard |
| 7.31919 | 0.292835 | standard |
| 7.33131 | 0.271423 | standard |
| 2.14252 | 0.999737 | standard |
| 3.82978 | 1.0 | standard |
| 7.00308 | 0.272163 | standard |
| 7.01451 | 0.292258 | standard |
| 7.31949 | 0.292774 | standard |
| 7.33091 | 0.272167 | standard |
| 2.14252 | 0.998834 | standard |
| 3.82978 | 1.0 | standard |
| 7.00377 | 0.273861 | standard |
| 7.01392 | 0.291771 | standard |
| 7.32018 | 0.291237 | standard |
| 7.33033 | 0.27209 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.999285 | standard |
| 7.00407 | 0.274429 | standard |
| 7.01362 | 0.291384 | standard |
| 7.32037 | 0.289866 | standard |
| 7.33003 | 0.27298 | standard |
| 2.14252 | 1.0 | standard |
| 3.82978 | 0.999555 | standard |
| 7.00426 | 0.274081 | standard |
| 7.01343 | 0.289554 | standard |
| 7.32068 | 0.291848 | standard |
| 7.32973 | 0.275689 | standard |
| 2.14252 | 0.999832 | standard |
| 3.82978 | 1.0 | standard |
| 7.00535 | 0.279963 | standard |
| 7.01244 | 0.284399 | standard |
| 7.32165 | 0.284479 | standard |
| 7.32864 | 0.282231 | standard |