2-Pyridinylmethanol
Simulation outputs:
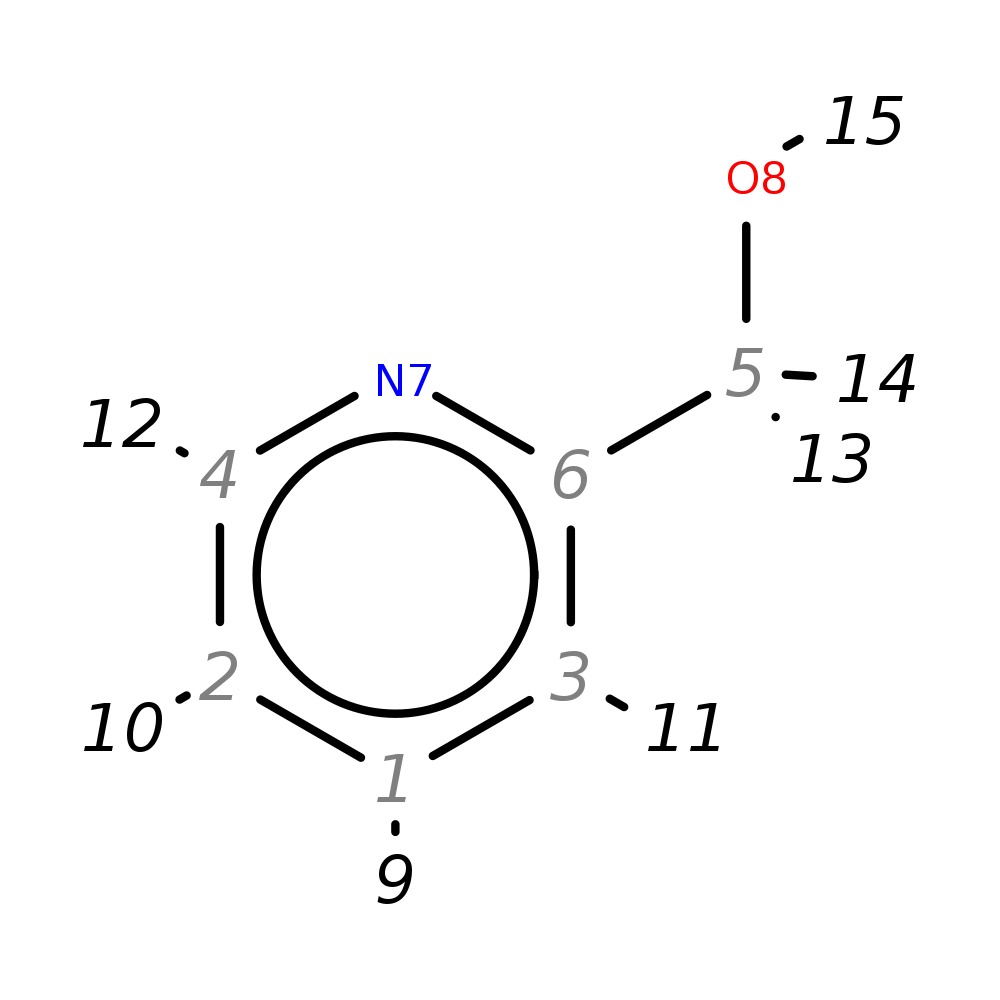
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.11736 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H7NO/c8-5-6-3-1-2-4-7-6/h1-4,8H,5H2 | |
| Note 1 | 13,14 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-Pyridinylmethanol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|
| 9 | 7.898 | 7.827 | 9.894 | 0 | 0 | 0 |
| 10 | 0 | 7.379 | 0 | 5.28 | 0 | 0 |
| 11 | 0 | 0 | 7.518 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 8.477 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 4.518 | 11.8 |
| 14 | 0 | 0 | 0 | 0 | 0 | 4.518 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.51793 | 1.0 | standard |
| 7.36836 | 0.130673 | standard |
| 7.37715 | 0.146874 | standard |
| 7.38127 | 0.149583 | standard |
| 7.39015 | 0.133756 | standard |
| 7.50959 | 0.241958 | standard |
| 7.52605 | 0.263396 | standard |
| 7.88353 | 0.135829 | standard |
| 7.89675 | 0.156522 | standard |
| 7.89983 | 0.150429 | standard |
| 7.91307 | 0.121844 | standard |
| 8.473 | 0.258644 | standard |
| 8.4818 | 0.258643 | standard |
| 4.51793 | 1.0 | standard |
| 7.1968 | 0.0739291 | standard |
| 7.30467 | 0.098232 | standard |
| 7.3867 | 0.264122 | standard |
| 7.51708 | 0.206817 | standard |
| 7.59742 | 0.375794 | standard |
| 7.74206 | 0.285316 | standard |
| 7.87236 | 0.0705781 | standard |
| 7.92632 | 0.103244 | standard |
| 8.16213 | 0.052911 | standard |
| 8.42146 | 0.26337 | standard |
| 8.54298 | 0.207722 | standard |
| 4.51793 | 1.0 | standard |
| 7.26262 | 0.094342 | standard |
| 7.34858 | 0.127334 | standard |
| 7.3902 | 0.18865 | standard |
| 7.42231 | 0.20108 | standard |
| 7.47458 | 0.189792 | standard |
| 7.58194 | 0.346206 | standard |
| 7.7794 | 0.229809 | standard |
| 7.9063 | 0.142125 | standard |
| 7.93878 | 0.112645 | standard |
| 8.06683 | 0.067187 | standard |
| 8.43648 | 0.277773 | standard |
| 8.52184 | 0.238517 | standard |
| 4.51793 | 1.0 | standard |
| 7.2931 | 0.104109 | standard |
| 7.35896 | 0.134914 | standard |
| 7.38835 | 0.167262 | standard |
| 7.45064 | 0.319137 | standard |
| 7.56953 | 0.328729 | standard |
| 7.80345 | 0.204083 | standard |
| 7.89969 | 0.151668 | standard |
| 7.92452 | 0.125839 | standard |
| 8.02136 | 0.0783692 | standard |
| 8.44586 | 0.273766 | standard |
| 8.51085 | 0.245208 | standard |
| 4.51793 | 1.0 | standard |
| 7.30307 | 0.107191 | standard |
| 7.36177 | 0.136807 | standard |
| 7.38798 | 0.164019 | standard |
| 7.45219 | 0.291307 | standard |
| 7.56485 | 0.321892 | standard |
| 7.81226 | 0.19446 | standard |
| 7.89822 | 0.153399 | standard |
| 7.92018 | 0.129928 | standard |
| 8.0066 | 0.082621 | standard |
| 8.44917 | 0.272125 | standard |
| 8.50712 | 0.246713 | standard |
| 4.51793 | 1.0 | standard |
| 7.31096 | 0.109699 | standard |
| 7.36391 | 0.138098 | standard |
| 7.38763 | 0.162087 | standard |
| 7.44123 | 0.191355 | standard |
| 7.46222 | 0.218953 | standard |
| 7.56093 | 0.315685 | standard |
| 7.81967 | 0.186734 | standard |
| 7.89724 | 0.154366 | standard |
| 7.91695 | 0.132963 | standard |
| 7.99491 | 0.086236 | standard |
| 8.45184 | 0.270773 | standard |
| 8.50416 | 0.247829 | standard |
| 4.51793 | 1.0 | standard |
| 7.34578 | 0.122115 | standard |
| 7.37238 | 0.141624 | standard |
| 7.38452 | 0.156297 | standard |
| 7.41106 | 0.14501 | standard |
| 7.49174 | 0.221787 | standard |
| 7.54112 | 0.285597 | standard |
| 7.85621 | 0.156242 | standard |
| 7.89553 | 0.155794 | standard |
| 7.90513 | 0.145051 | standard |
| 7.94455 | 0.105188 | standard |
| 8.46439 | 0.261723 | standard |
| 8.49068 | 0.255916 | standard |
| 4.51793 | 1.0 | standard |
| 7.35712 | 0.126156 | standard |
| 7.37486 | 0.143123 | standard |
| 7.38305 | 0.154191 | standard |
| 7.4007 | 0.13914 | standard |
| 7.50081 | 0.231811 | standard |
| 7.53379 | 0.27448 | standard |
| 7.86954 | 0.14663 | standard |
| 7.89587 | 0.155156 | standard |
| 7.90227 | 0.147855 | standard |
| 7.92854 | 0.112518 | standard |
| 8.46864 | 0.258642 | standard |
| 8.48625 | 0.258641 | standard |
| 4.51793 | 1.0 | standard |
| 7.36277 | 0.12754 | standard |
| 7.37606 | 0.144375 | standard |
| 7.38218 | 0.152662 | standard |
| 7.39545 | 0.137175 | standard |
| 7.50528 | 0.236747 | standard |
| 7.53002 | 0.268918 | standard |
| 7.87649 | 0.141999 | standard |
| 7.89621 | 0.155048 | standard |
| 7.90096 | 0.149553 | standard |
| 7.92071 | 0.116371 | standard |
| 8.47082 | 0.258693 | standard |
| 8.48398 | 0.258693 | standard |
| 4.51793 | 1.0 | standard |
| 7.36608 | 0.128389 | standard |
| 7.37677 | 0.14527 | standard |
| 7.38165 | 0.151941 | standard |
| 7.39223 | 0.136178 | standard |
| 7.5079 | 0.239914 | standard |
| 7.52764 | 0.265616 | standard |
| 7.88075 | 0.139278 | standard |
| 7.89647 | 0.154304 | standard |
| 7.90031 | 0.149861 | standard |
| 7.91615 | 0.118727 | standard |
| 8.4721 | 0.258595 | standard |
| 8.48269 | 0.258594 | standard |
| 4.51793 | 1.0 | standard |
| 7.36836 | 0.130673 | standard |
| 7.37715 | 0.146875 | standard |
| 7.38127 | 0.149584 | standard |
| 7.39015 | 0.133757 | standard |
| 7.50959 | 0.241958 | standard |
| 7.52605 | 0.263396 | standard |
| 7.88353 | 0.135829 | standard |
| 7.89675 | 0.156523 | standard |
| 7.89983 | 0.15043 | standard |
| 7.91307 | 0.121844 | standard |
| 8.473 | 0.258644 | standard |
| 8.4818 | 0.258644 | standard |
| 4.51793 | 1.0 | standard |
| 7.36995 | 0.130825 | standard |
| 7.37754 | 0.147754 | standard |
| 7.38098 | 0.150109 | standard |
| 7.38857 | 0.133569 | standard |
| 7.51088 | 0.243458 | standard |
| 7.52496 | 0.261842 | standard |
| 7.88561 | 0.134832 | standard |
| 7.89682 | 0.154966 | standard |
| 7.89957 | 0.149649 | standard |
| 7.91089 | 0.122789 | standard |
| 8.47359 | 0.2587 | standard |
| 8.48111 | 0.2587 | standard |
| 4.51793 | 1.0 | standard |
| 7.37054 | 0.131489 | standard |
| 7.37763 | 0.147971 | standard |
| 7.38088 | 0.14911 | standard |
| 7.38797 | 0.133427 | standard |
| 7.51133 | 0.243708 | standard |
| 7.52446 | 0.261202 | standard |
| 7.8864 | 0.134387 | standard |
| 7.89691 | 0.154684 | standard |
| 7.89947 | 0.14926 | standard |
| 7.91 | 0.124401 | standard |
| 8.47389 | 0.258675 | standard |
| 8.48091 | 0.258675 | standard |
| 4.51793 | 1.0 | standard |
| 7.37114 | 0.132207 | standard |
| 7.37773 | 0.148438 | standard |
| 7.38079 | 0.148359 | standard |
| 7.38748 | 0.13205 | standard |
| 7.51177 | 0.244562 | standard |
| 7.52407 | 0.260651 | standard |
| 7.8871 | 0.132791 | standard |
| 7.89701 | 0.155989 | standard |
| 7.89938 | 0.149541 | standard |
| 7.90921 | 0.124696 | standard |
| 8.47408 | 0.25856 | standard |
| 8.48071 | 0.25856 | standard |
| 4.51793 | 1.0 | standard |
| 7.37203 | 0.132108 | standard |
| 7.37791 | 0.147814 | standard |
| 7.3807 | 0.147816 | standard |
| 7.38659 | 0.132115 | standard |
| 7.51247 | 0.245408 | standard |
| 7.52347 | 0.259715 | standard |
| 7.88829 | 0.132325 | standard |
| 7.8971 | 0.155815 | standard |
| 7.89919 | 0.150083 | standard |
| 7.90802 | 0.125075 | standard |
| 8.47448 | 0.258716 | standard |
| 8.48032 | 0.258716 | standard |
| 4.51793 | 1.0 | standard |
| 7.37242 | 0.132122 | standard |
| 7.37801 | 0.148346 | standard |
| 7.3806 | 0.148347 | standard |
| 7.38619 | 0.132128 | standard |
| 7.51276 | 0.249201 | standard |
| 7.52318 | 0.255978 | standard |
| 7.88878 | 0.13043 | standard |
| 7.89709 | 0.153965 | standard |
| 7.8991 | 0.151235 | standard |
| 7.90742 | 0.126995 | standard |
| 8.47458 | 0.258708 | standard |
| 8.48012 | 0.258709 | standard |
| 4.51793 | 1.0 | standard |
| 7.37272 | 0.131988 | standard |
| 7.37811 | 0.149059 | standard |
| 7.3805 | 0.14916 | standard |
| 7.38579 | 0.132199 | standard |
| 7.51306 | 0.249404 | standard |
| 7.52288 | 0.255841 | standard |
| 7.88928 | 0.130325 | standard |
| 7.89719 | 0.153684 | standard |
| 7.89911 | 0.151084 | standard |
| 7.90702 | 0.127062 | standard |
| 8.47478 | 0.258769 | standard |
| 8.48002 | 0.25877 | standard |
| 4.51793 | 1.0 | standard |
| 7.3743 | 0.132135 | standard |
| 7.3784 | 0.149529 | standard |
| 7.38021 | 0.149529 | standard |
| 7.38431 | 0.132137 | standard |
| 7.51415 | 0.251295 | standard |
| 7.52179 | 0.253791 | standard |
| 7.89126 | 0.129901 | standard |
| 7.89737 | 0.152506 | standard |
| 7.89883 | 0.151509 | standard |
| 7.90494 | 0.12863 | standard |
| 8.47537 | 0.258668 | standard |
| 8.47943 | 0.258656 | standard |