7-Methyl-2,3-dihydro-1H-inden-4-yl acetate
Simulation outputs:
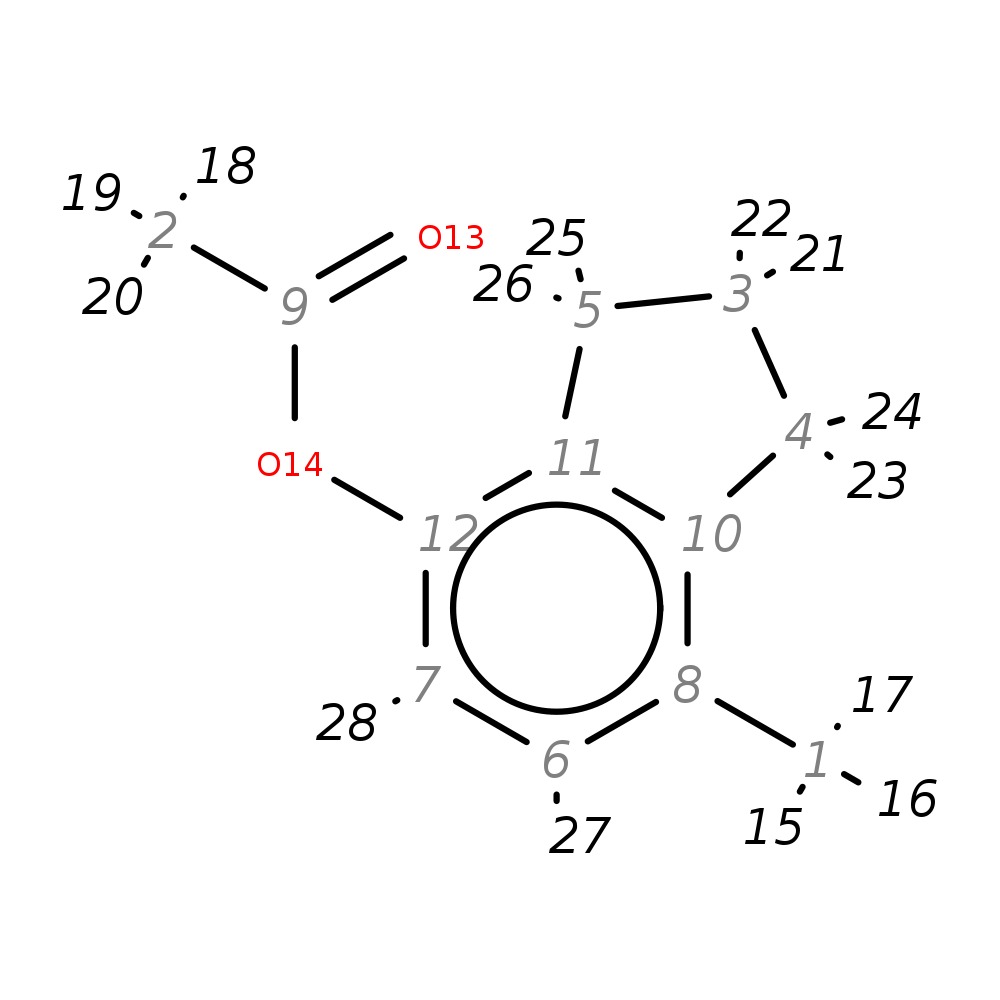
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.06278 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H14O2/c1-8-6-7-12(14-9(2)13)11-5-3-4-10(8)11/h6-7H,3-5H2,1-2H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 7-Methyl-2,3-dihydro-1H-inden-4-yl acetate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 2.33 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 2.33 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 2.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 2.245 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 2.245 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 2.245 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 2.064 | -14.0 | 7.873 | 7.873 | 8.024 | 8.024 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.064 | 7.873 | 7.873 | 8.024 | 8.024 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.76 | -14.0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.76 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.877 | -14.0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.877 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.826 | 8.076 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.07 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.03691 | 0.0436616 | standard |
| 2.0501 | 0.169383 | standard |
| 2.06335 | 0.251434 | standard |
| 2.07658 | 0.169457 | standard |
| 2.08982 | 0.0465973 | standard |
| 2.24517 | 1.0 | standard |
| 2.32958 | 0.999045 | standard |
| 2.74677 | 0.173512 | standard |
| 2.75987 | 0.336532 | standard |
| 2.77299 | 0.169735 | standard |
| 2.86341 | 0.173131 | standard |
| 2.87679 | 0.336878 | standard |
| 2.89018 | 0.170347 | standard |
| 6.81945 | 0.161472 | standard |
| 6.83284 | 0.175973 | standard |
| 7.06375 | 0.175815 | standard |
| 7.07723 | 0.161745 | standard |
| 1.59159 | 0.0112421 | standard |
| 1.67629 | 0.0153761 | standard |
| 1.86623 | 0.0529431 | standard |
| 2.05459 | 0.117311 | standard |
| 2.24532 | 1.0 | standard |
| 2.32933 | 0.951568 | standard |
| 2.59381 | 0.193954 | standard |
| 2.75871 | 0.274255 | standard |
| 2.88041 | 0.224898 | standard |
| 3.07085 | 0.0636682 | standard |
| 6.68927 | 0.0536471 | standard |
| 6.8912 | 0.237451 | standard |
| 7.00549 | 0.237785 | standard |
| 7.20742 | 0.0542623 | standard |
| 1.76204 | 0.015707 | standard |
| 1.80256 | 0.0188461 | standard |
| 1.91573 | 0.0743861 | standard |
| 2.03976 | 0.160081 | standard |
| 2.24533 | 0.995313 | standard |
| 2.32925 | 1.0 | standard |
| 2.6405 | 0.187892 | standard |
| 2.763 | 0.407362 | standard |
| 2.88345 | 0.310513 | standard |
| 3.01025 | 0.0868585 | standard |
| 6.74179 | 0.084061 | standard |
| 6.8763 | 0.235459 | standard |
| 7.02038 | 0.23558 | standard |
| 7.1549 | 0.084464 | standard |
| 1.84402 | 0.0185132 | standard |
| 1.86628 | 0.020656 | standard |
| 1.95518 | 0.0905542 | standard |
| 2.0505 | 0.185259 | standard |
| 2.14634 | 0.169862 | standard |
| 2.24523 | 1.00001 | standard |
| 2.32954 | 0.962447 | standard |
| 2.66878 | 0.180582 | standard |
| 2.77485 | 0.327246 | standard |
| 2.87798 | 0.309986 | standard |
| 2.97975 | 0.102972 | standard |
| 6.76584 | 0.0997364 | standard |
| 6.86676 | 0.218891 | standard |
| 7.02993 | 0.21902 | standard |
| 7.13075 | 0.0998106 | standard |
| 1.87065 | 0.020127 | standard |
| 1.88766 | 0.0219302 | standard |
| 1.96784 | 0.098989 | standard |
| 2.05332 | 0.196787 | standard |
| 2.13876 | 0.170466 | standard |
| 2.24507 | 1.0 | standard |
| 2.32956 | 0.97339 | standard |
| 2.67838 | 0.182264 | standard |
| 2.76453 | 0.287676 | standard |
| 2.79189 | 0.231743 | standard |
| 2.87967 | 0.290172 | standard |
| 2.96899 | 0.112002 | standard |
| 6.77347 | 0.107087 | standard |
| 6.86318 | 0.216101 | standard |
| 7.0335 | 0.216466 | standard |
| 7.12311 | 0.107461 | standard |
| 1.89182 | 0.0218403 | standard |
| 1.90472 | 0.0233501 | standard |
| 1.97779 | 0.106899 | standard |
| 2.05532 | 0.207578 | standard |
| 2.13263 | 0.17335 | standard |
| 2.24517 | 1.0 | standard |
| 2.32957 | 0.992256 | standard |
| 2.68601 | 0.185221 | standard |
| 2.76346 | 0.291376 | standard |
| 2.80127 | 0.214186 | standard |
| 2.84203 | 0.146888 | standard |
| 2.87975 | 0.296518 | standard |
| 2.95983 | 0.120484 | standard |
| 6.77951 | 0.114838 | standard |
| 6.86021 | 0.216357 | standard |
| 7.03648 | 0.216131 | standard |
| 7.11717 | 0.114862 | standard |
| 1.98247 | 0.0329846 | standard |
| 2.02195 | 0.141508 | standard |
| 2.06149 | 0.238386 | standard |
| 2.10107 | 0.17678 | standard |
| 2.14068 | 0.0508936 | standard |
| 2.24518 | 1.0 | standard |
| 2.32958 | 0.999384 | standard |
| 2.72159 | 0.18418 | standard |
| 2.76085 | 0.321699 | standard |
| 2.80009 | 0.151642 | standard |
| 2.83769 | 0.186321 | standard |
| 2.87766 | 0.324939 | standard |
| 2.91767 | 0.150496 | standard |
| 6.80449 | 0.141028 | standard |
| 6.84483 | 0.194842 | standard |
| 7.05185 | 0.19486 | standard |
| 7.0922 | 0.141791 | standard |
| 2.00982 | 0.0375334 | standard |
| 2.03623 | 0.153616 | standard |
| 2.06267 | 0.246461 | standard |
| 2.0891 | 0.176806 | standard |
| 2.11555 | 0.0493192 | standard |
| 2.24517 | 0.999648 | standard |
| 2.32958 | 1.0 | standard |
| 2.73405 | 0.181979 | standard |
| 2.76025 | 0.330557 | standard |
| 2.78644 | 0.157626 | standard |
| 2.85041 | 0.181299 | standard |
| 2.87711 | 0.332394 | standard |
| 2.90382 | 0.158881 | standard |
| 6.81212 | 0.150699 | standard |
| 6.83908 | 0.185997 | standard |
| 7.0576 | 0.186374 | standard |
| 7.08457 | 0.150819 | standard |
| 2.02338 | 0.0397298 | standard |
| 2.04321 | 0.162373 | standard |
| 2.06305 | 0.248993 | standard |
| 2.08291 | 0.173749 | standard |
| 2.10276 | 0.0485066 | standard |
| 2.24517 | 0.999949 | standard |
| 2.32958 | 1.0 | standard |
| 2.74037 | 0.180112 | standard |
| 2.76003 | 0.333432 | standard |
| 2.77967 | 0.163342 | standard |
| 2.85691 | 0.176956 | standard |
| 2.87694 | 0.334296 | standard |
| 2.89696 | 0.164347 | standard |
| 6.81579 | 0.155622 | standard |
| 6.83601 | 0.181654 | standard |
| 7.06067 | 0.181731 | standard |
| 7.0808 | 0.155827 | standard |
| 2.03151 | 0.0418634 | standard |
| 2.04739 | 0.166207 | standard |
| 2.06325 | 0.250856 | standard |
| 2.07913 | 0.170136 | standard |
| 2.09504 | 0.0481319 | standard |
| 2.24517 | 0.999436 | standard |
| 2.32958 | 1.0 | standard |
| 2.74419 | 0.176369 | standard |
| 2.75993 | 0.335194 | standard |
| 2.77566 | 0.166067 | standard |
| 2.86078 | 0.17381 | standard |
| 2.87684 | 0.336008 | standard |
| 2.89288 | 0.169216 | standard |
| 6.81797 | 0.158575 | standard |
| 6.83413 | 0.178429 | standard |
| 7.06256 | 0.178894 | standard |
| 7.07872 | 0.158309 | standard |
| 2.03691 | 0.0436776 | standard |
| 2.0501 | 0.169444 | standard |
| 2.06335 | 0.251489 | standard |
| 2.07658 | 0.169517 | standard |
| 2.08982 | 0.0466143 | standard |
| 2.24517 | 1.0 | standard |
| 2.32958 | 0.999839 | standard |
| 2.74677 | 0.173576 | standard |
| 2.75987 | 0.33672 | standard |
| 2.77299 | 0.169798 | standard |
| 2.86341 | 0.173194 | standard |
| 2.87679 | 0.337029 | standard |
| 2.89018 | 0.170409 | standard |
| 6.81945 | 0.161548 | standard |
| 6.83284 | 0.176004 | standard |
| 7.06375 | 0.175887 | standard |
| 7.07723 | 0.161813 | standard |
| 2.04072 | 0.0445602 | standard |
| 2.05206 | 0.16964 | standard |
| 2.06341 | 0.250393 | standard |
| 2.07477 | 0.169691 | standard |
| 2.08611 | 0.0465351 | standard |
| 2.24517 | 1.0 | standard |
| 2.32958 | 0.999603 | standard |
| 2.7486 | 0.173878 | standard |
| 2.75985 | 0.337195 | standard |
| 2.77105 | 0.170696 | standard |
| 2.86529 | 0.172336 | standard |
| 2.87675 | 0.336847 | standard |
| 2.88819 | 0.171125 | standard |
| 6.82044 | 0.163263 | standard |
| 6.83194 | 0.174363 | standard |
| 7.06474 | 0.174379 | standard |
| 7.07624 | 0.16314 | standard |
| 2.04231 | 0.0445148 | standard |
| 2.05283 | 0.169807 | standard |
| 2.06345 | 0.252348 | standard |
| 2.07402 | 0.169783 | standard |
| 2.08462 | 0.0457453 | standard |
| 2.24517 | 0.999646 | standard |
| 2.32958 | 1.0 | standard |
| 2.74934 | 0.172839 | standard |
| 2.75985 | 0.336611 | standard |
| 2.77031 | 0.171102 | standard |
| 2.86604 | 0.172075 | standard |
| 2.87674 | 0.337684 | standard |
| 2.88745 | 0.172043 | standard |
| 6.82084 | 0.164251 | standard |
| 6.83155 | 0.173409 | standard |
| 7.06504 | 0.172987 | standard |
| 7.07584 | 0.164022 | standard |
| 2.04359 | 0.0450639 | standard |
| 2.05353 | 0.169797 | standard |
| 2.06346 | 0.251325 | standard |
| 2.07338 | 0.170488 | standard |
| 2.08334 | 0.0456719 | standard |
| 2.24517 | 1.0 | standard |
| 2.32958 | 0.999819 | standard |
| 2.74999 | 0.173098 | standard |
| 2.7598 | 0.337094 | standard |
| 2.76967 | 0.171109 | standard |
| 2.86673 | 0.172314 | standard |
| 2.87674 | 0.338113 | standard |
| 2.88676 | 0.172288 | standard |
| 6.82114 | 0.163984 | standard |
| 6.83125 | 0.173156 | standard |
| 7.06543 | 0.173375 | standard |
| 7.07545 | 0.164764 | standard |
| 2.04582 | 0.045254 | standard |
| 2.05464 | 0.169837 | standard |
| 2.06347 | 0.252531 | standard |
| 2.07229 | 0.169843 | standard |
| 2.08111 | 0.045275 | standard |
| 2.24517 | 0.998537 | standard |
| 2.32958 | 1.0 | standard |
| 2.75108 | 0.172182 | standard |
| 2.7598 | 0.337635 | standard |
| 2.76853 | 0.1722 | standard |
| 2.86782 | 0.17194 | standard |
| 2.87674 | 0.33741 | standard |
| 2.88567 | 0.171921 | standard |
| 6.82173 | 0.165367 | standard |
| 6.83075 | 0.172384 | standard |
| 7.06593 | 0.172103 | standard |
| 7.07485 | 0.165717 | standard |
| 2.04681 | 0.0452211 | standard |
| 2.05514 | 0.169574 | standard |
| 2.06348 | 0.252681 | standard |
| 2.07187 | 0.170315 | standard |
| 2.08022 | 0.0452121 | standard |
| 2.24517 | 0.999481 | standard |
| 2.32958 | 1.0 | standard |
| 2.75148 | 0.171979 | standard |
| 2.7598 | 0.337336 | standard |
| 2.76813 | 0.171995 | standard |
| 2.86832 | 0.171857 | standard |
| 2.87674 | 0.33722 | standard |
| 2.88517 | 0.171842 | standard |
| 6.82203 | 0.165192 | standard |
| 6.83046 | 0.171585 | standard |
| 7.06613 | 0.171807 | standard |
| 7.07465 | 0.164949 | standard |
| 2.04761 | 0.0452 | standard |
| 2.05556 | 0.170424 | standard |
| 2.0635 | 0.253337 | standard |
| 2.07145 | 0.170297 | standard |
| 2.07942 | 0.045187 | standard |
| 2.24517 | 1.0 | standard |
| 2.32958 | 0.999895 | standard |
| 2.75196 | 0.172293 | standard |
| 2.7598 | 0.337793 | standard |
| 2.76763 | 0.172294 | standard |
| 2.86871 | 0.172002 | standard |
| 2.87674 | 0.337166 | standard |
| 2.88472 | 0.172468 | standard |
| 6.82223 | 0.1669 | standard |
| 6.83026 | 0.170773 | standard |
| 7.06633 | 0.170681 | standard |
| 7.07446 | 0.167146 | standard |
| 2.05132 | 0.0453998 | standard |
| 2.05744 | 0.170683 | standard |
| 2.06355 | 0.253498 | standard |
| 2.06964 | 0.170697 | standard |
| 2.07581 | 0.0453586 | standard |
| 2.24517 | 0.999864 | standard |
| 2.32958 | 1.0 | standard |
| 2.75371 | 0.172495 | standard |
| 2.7598 | 0.337025 | standard |
| 2.76585 | 0.171873 | standard |
| 2.87055 | 0.172401 | standard |
| 2.87669 | 0.338327 | standard |
| 2.88284 | 0.172395 | standard |
| 6.82322 | 0.168465 | standard |
| 6.82937 | 0.16944 | standard |
| 7.06722 | 0.169929 | standard |
| 7.07346 | 0.168012 | standard |