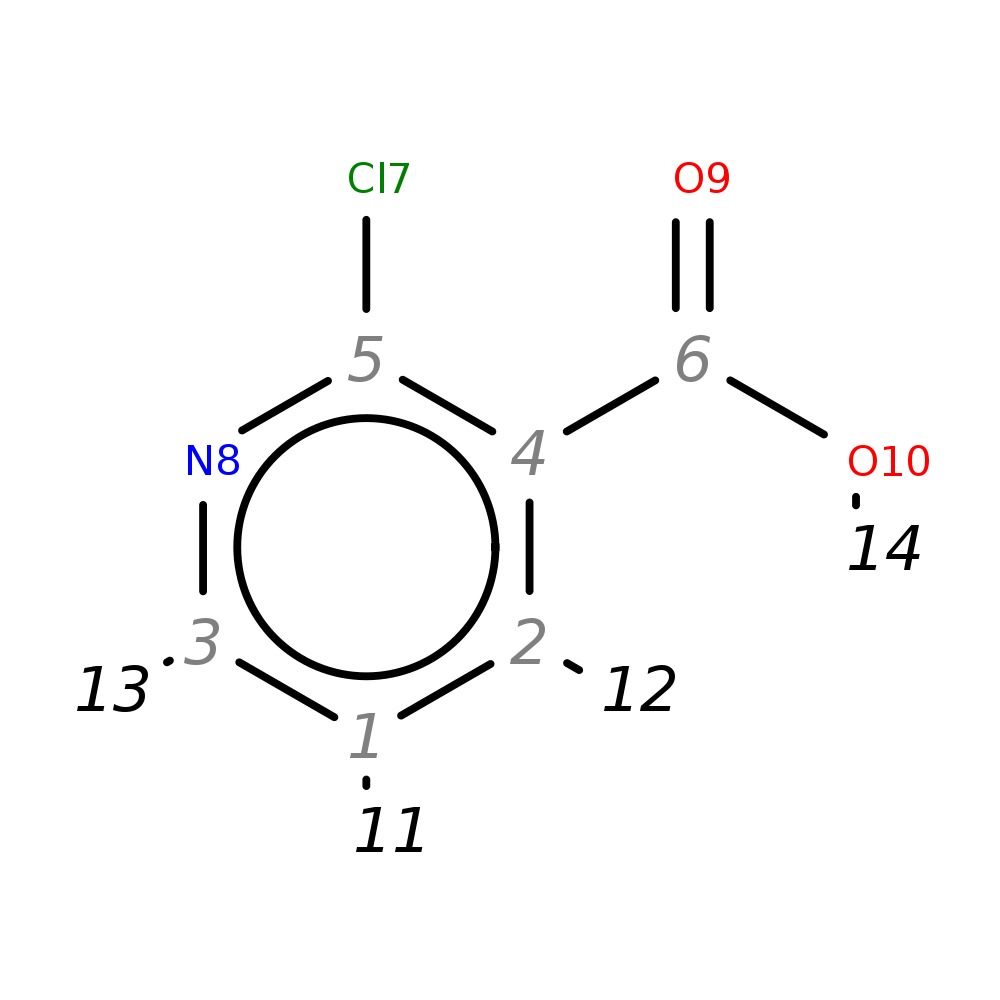
2-Chloronicotinic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02908 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H4ClNO2/c7-5-4(6(9)10)2-1-3-8-5/h1-3H,(H,9,10) | |
| Note 1 | 12?13 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-Chloronicotinic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | |
|---|---|---|---|
| 11 | 7.44 | 8.233 | 4.823 |
| 12 | 0 | 7.865 | 1.606 |
| 13 | 0 | 0 | 8.332 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 7.42877 | 0.725174 | standard |
| 7.43681 | 0.778843 | standard |
| 7.44242 | 0.821817 | standard |
| 7.45046 | 0.771579 | standard |
| 7.85711 | 0.96183 | standard |
| 7.85926 | 0.968633 | standard |
| 7.87078 | 0.911238 | standard |
| 7.87293 | 0.903992 | standard |
| 8.32689 | 0.963498 | standard |
| 8.32907 | 1.0 | standard |
| 8.33482 | 1.0 | standard |
| 8.337 | 0.963497 | standard |
| 7.25225 | 0.292333 | standard |
| 7.36724 | 0.375806 | standard |
| 7.45803 | 0.687064 | standard |
| 7.57225 | 0.886818 | standard |
| 7.76559 | 0.852212 | standard |
| 7.80555 | 1.0 | standard |
| 7.96984 | 0.400534 | standard |
| 8.01049 | 0.378957 | standard |
| 8.25909 | 0.786569 | standard |
| 8.30031 | 0.7342 | standard |
| 8.37205 | 0.656143 | standard |
| 8.41308 | 0.562581 | standard |
| 7.31949 | 0.399084 | standard |
| 7.39817 | 0.49482 | standard |
| 7.45659 | 0.738538 | standard |
| 7.53485 | 0.8548 | standard |
| 7.7952 | 0.907385 | standard |
| 7.8193 | 1.00001 | standard |
| 7.93169 | 0.525471 | standard |
| 7.95602 | 0.521198 | standard |
| 8.28278 | 0.845269 | standard |
| 8.30733 | 0.823119 | standard |
| 8.36013 | 0.753994 | standard |
| 8.38472 | 0.677188 | standard |
| 7.35155 | 0.470548 | standard |
| 7.41121 | 0.567923 | standard |
| 7.45427 | 0.765914 | standard |
| 7.51365 | 0.83911 | standard |
| 7.81107 | 0.933803 | standard |
| 7.82838 | 1.0 | standard |
| 7.91363 | 0.613149 | standard |
| 7.93091 | 0.615217 | standard |
| 8.29484 | 0.881935 | standard |
| 8.31239 | 0.874101 | standard |
| 8.3535 | 0.815021 | standard |
| 8.37112 | 0.746947 | standard |
| 7.36204 | 0.496927 | standard |
| 7.41513 | 0.593294 | standard |
| 7.45328 | 0.773722 | standard |
| 7.50622 | 0.832372 | standard |
| 7.81662 | 0.942539 | standard |
| 7.83165 | 1.0 | standard |
| 7.90771 | 0.64415 | standard |
| 7.92297 | 0.647604 | standard |
| 8.29885 | 0.892445 | standard |
| 8.31435 | 0.889607 | standard |
| 8.35116 | 0.836368 | standard |
| 8.36659 | 0.771262 | standard |
| 7.37026 | 0.520503 | standard |
| 7.41807 | 0.615563 | standard |
| 7.45238 | 0.781615 | standard |
| 7.50018 | 0.828883 | standard |
| 7.82107 | 0.948651 | standard |
| 7.83454 | 1.0 | standard |
| 7.90313 | 0.67341 | standard |
| 7.91666 | 0.677655 | standard |
| 8.30211 | 0.905077 | standard |
| 8.31585 | 0.905885 | standard |
| 8.34932 | 0.856292 | standard |
| 8.36301 | 0.793905 | standard |
| 7.4061 | 0.645872 | standard |
| 7.43014 | 0.729456 | standard |
| 7.44714 | 0.82159 | standard |
| 7.47115 | 0.816727 | standard |
| 7.84209 | 0.991493 | standard |
| 7.84854 | 1.00002 | standard |
| 7.88317 | 0.833089 | standard |
| 7.88964 | 0.823365 | standard |
| 8.31693 | 0.956632 | standard |
| 8.32364 | 0.993332 | standard |
| 8.34074 | 0.953647 | standard |
| 8.34731 | 0.9156 | standard |
| 7.41758 | 0.702887 | standard |
| 7.43357 | 0.756176 | standard |
| 7.44488 | 0.841858 | standard |
| 7.46095 | 0.795177 | standard |
| 7.84942 | 0.983705 | standard |
| 7.85388 | 0.991115 | standard |
| 7.87688 | 0.882983 | standard |
| 7.88119 | 0.874909 | standard |
| 8.32193 | 0.963504 | standard |
| 8.3263 | 0.999999 | standard |
| 8.33779 | 1.0 | standard |
| 8.34216 | 0.963496 | standard |
| 7.42322 | 0.711271 | standard |
| 7.43524 | 0.764425 | standard |
| 7.4437 | 0.828304 | standard |
| 7.4558 | 0.780022 | standard |
| 7.85329 | 0.976908 | standard |
| 7.85644 | 0.983633 | standard |
| 7.87377 | 0.889661 | standard |
| 7.87707 | 0.88196 | standard |
| 8.32447 | 0.96441 | standard |
| 8.32767 | 1.0 | standard |
| 8.33629 | 0.992429 | standard |
| 8.33964 | 0.955309 | standard |
| 7.42659 | 0.718066 | standard |
| 7.43622 | 0.770792 | standard |
| 7.44301 | 0.822249 | standard |
| 7.45263 | 0.773554 | standard |
| 7.85559 | 0.968529 | standard |
| 7.85811 | 0.975138 | standard |
| 7.87203 | 0.906175 | standard |
| 7.87455 | 0.899054 | standard |
| 8.32597 | 0.964669 | standard |
| 8.32853 | 1.00026 | standard |
| 8.33546 | 1.00026 | standard |
| 8.33802 | 0.964667 | standard |
| 7.42877 | 0.725169 | standard |
| 7.43681 | 0.778839 | standard |
| 7.44242 | 0.821814 | standard |
| 7.45046 | 0.771574 | standard |
| 7.85711 | 0.961829 | standard |
| 7.85926 | 0.968632 | standard |
| 7.87078 | 0.911235 | standard |
| 7.87293 | 0.903991 | standard |
| 8.32689 | 0.963498 | standard |
| 8.32907 | 1.0 | standard |
| 8.33482 | 1.0 | standard |
| 8.337 | 0.963497 | standard |
| 7.43035 | 0.728733 | standard |
| 7.4372 | 0.779819 | standard |
| 7.44212 | 0.816828 | standard |
| 7.44897 | 0.768546 | standard |
| 7.85814 | 0.945191 | standard |
| 7.86011 | 0.952393 | standard |
| 7.86996 | 0.917459 | standard |
| 7.87178 | 0.910399 | standard |
| 8.32764 | 0.965078 | standard |
| 8.32949 | 1.00015 | standard |
| 8.3345 | 1.00015 | standard |
| 8.33634 | 0.965077 | standard |
| 7.43095 | 0.733988 | standard |
| 7.4374 | 0.786877 | standard |
| 7.44192 | 0.821525 | standard |
| 7.44838 | 0.771349 | standard |
| 7.85864 | 0.955564 | standard |
| 7.86041 | 0.962567 | standard |
| 7.86966 | 0.932099 | standard |
| 7.87128 | 0.925334 | standard |
| 8.32791 | 0.962763 | standard |
| 8.32971 | 1.0007 | standard |
| 8.33427 | 1.0007 | standard |
| 8.33608 | 0.962763 | standard |
| 7.43154 | 0.729101 | standard |
| 7.4376 | 0.781811 | standard |
| 7.44183 | 0.814011 | standard |
| 7.44788 | 0.76384 | standard |
| 7.85904 | 0.957102 | standard |
| 7.86061 | 0.963571 | standard |
| 7.86934 | 0.920368 | standard |
| 7.87091 | 0.913582 | standard |
| 8.32821 | 0.965807 | standard |
| 8.3298 | 1.00058 | standard |
| 8.33418 | 1.00058 | standard |
| 8.33578 | 0.965806 | standard |
| 7.43243 | 0.732796 | standard |
| 7.43779 | 0.783442 | standard |
| 7.44163 | 0.81224 | standard |
| 7.44699 | 0.763765 | standard |
| 7.8597 | 0.952569 | standard |
| 7.86113 | 0.95931 | standard |
| 7.86881 | 0.92102 | standard |
| 7.87024 | 0.913985 | standard |
| 8.32857 | 0.963866 | standard |
| 8.33003 | 1.00044 | standard |
| 8.33386 | 1.00044 | standard |
| 8.33531 | 0.963866 | standard |
| 7.43283 | 0.732642 | standard |
| 7.43789 | 0.783002 | standard |
| 7.44153 | 0.810254 | standard |
| 7.44659 | 0.761941 | standard |
| 7.85999 | 0.951152 | standard |
| 7.86134 | 0.957875 | standard |
| 7.86861 | 0.921619 | standard |
| 7.86995 | 0.914617 | standard |
| 8.32876 | 0.963678 | standard |
| 8.33013 | 1.00033 | standard |
| 8.33375 | 1.00033 | standard |
| 8.33512 | 0.963678 | standard |
| 7.43323 | 0.747788 | standard |
| 7.43799 | 0.797602 | standard |
| 7.44143 | 0.797604 | standard |
| 7.4462 | 0.747788 | standard |
| 7.86019 | 0.935784 | standard |
| 7.86144 | 0.942337 | standard |
| 7.86841 | 0.942338 | standard |
| 7.86966 | 0.935783 | standard |
| 8.32896 | 0.965834 | standard |
| 8.33023 | 1.00036 | standard |
| 8.33375 | 1.00036 | standard |
| 8.33503 | 0.965833 | standard |
| 7.43471 | 0.738776 | standard |
| 7.43848 | 0.792052 | standard |
| 7.44104 | 0.793181 | standard |
| 7.44471 | 0.740565 | standard |
| 7.86135 | 0.936219 | standard |
| 7.86226 | 0.942509 | standard |
| 7.86756 | 0.915867 | standard |
| 7.86862 | 0.908591 | standard |
| 8.32959 | 0.937571 | standard |
| 8.33067 | 0.975555 | standard |
| 8.33334 | 1.00004 | standard |
| 8.33427 | 0.966182 | standard |