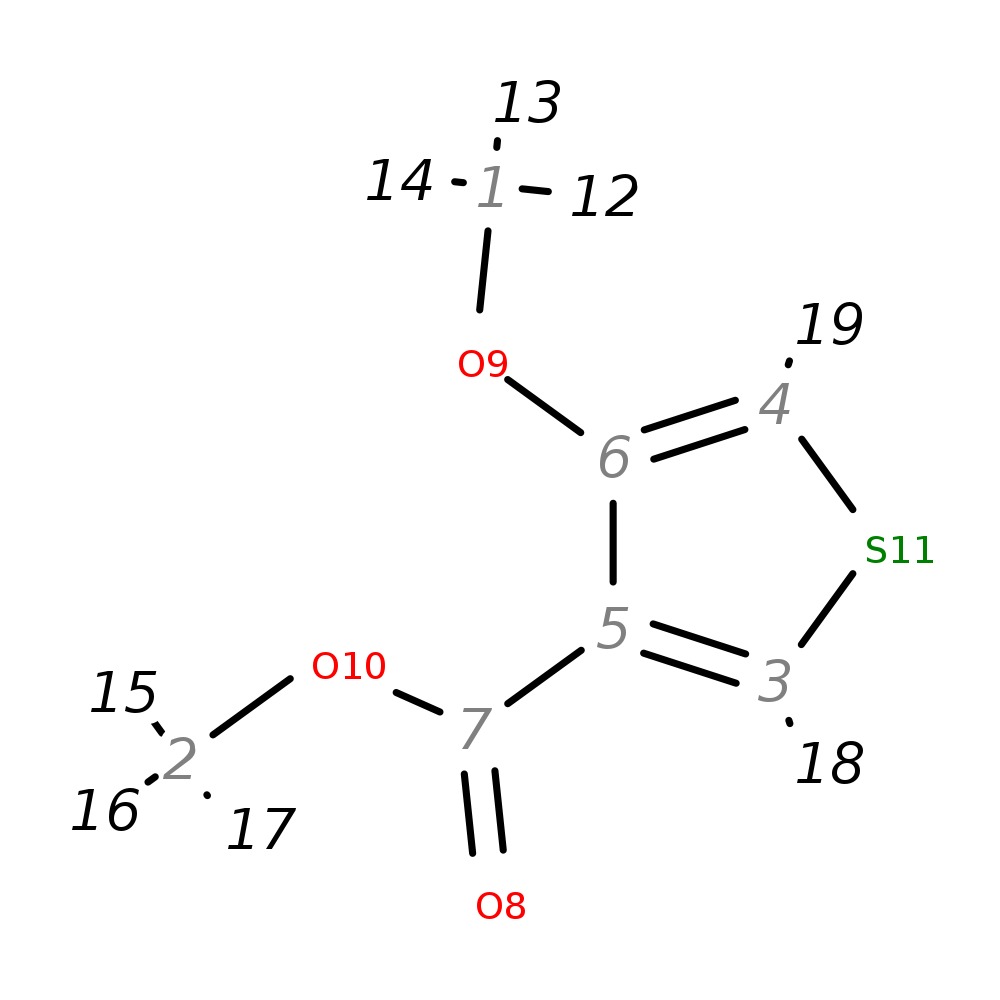
Methyl 4-methoxythiophene-3-carboxylate
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02357 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H8O3S/c1-9-6-4-11-3-5(6)7(8)10-2/h3-4H,1-2H3 | |
| Note 1 | 12,13,14?15,16,17 | |
| Note 2 | 18?19 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Methyl 4-methoxythiophene-3-carboxylate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|
| 12 | 3.888 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 3.888 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 3.888 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 3.847 | -14.0 | -14.0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 3.847 | -14.0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 3.847 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 8.225 | 2.892 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.648 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.84703 | 0.999562 | standard |
| 3.8876 | 1.0 | standard |
| 6.64603 | 0.183746 | standard |
| 6.65078 | 0.184007 | standard |
| 8.2222 | 0.184639 | standard |
| 8.22684 | 0.184218 | standard |
| 3.8509 | 0.999659 | standard |
| 3.88374 | 1.0 | standard |
| 6.61227 | 0.138974 | standard |
| 6.68287 | 0.146221 | standard |
| 8.19002 | 0.146295 | standard |
| 8.26061 | 0.138899 | standard |
| 3.84793 | 0.999905 | standard |
| 3.8867 | 1.0 | standard |
| 6.62453 | 0.159477 | standard |
| 6.6716 | 0.162126 | standard |
| 8.20137 | 0.162014 | standard |
| 8.24844 | 0.159729 | standard |
| 3.84735 | 0.99907 | standard |
| 3.88729 | 1.0 | standard |
| 6.63053 | 0.169331 | standard |
| 6.66578 | 0.169449 | standard |
| 8.2071 | 0.169585 | standard |
| 8.24235 | 0.169454 | standard |
| 3.84723 | 1.0 | standard |
| 3.8874 | 0.999178 | standard |
| 6.63256 | 0.172258 | standard |
| 6.66394 | 0.17215 | standard |
| 8.20903 | 0.172459 | standard |
| 8.24041 | 0.171828 | standard |
| 3.84717 | 0.999654 | standard |
| 3.88747 | 1.00001 | standard |
| 6.63411 | 0.174169 | standard |
| 6.66239 | 0.17444 | standard |
| 8.21058 | 0.174106 | standard |
| 8.23876 | 0.174508 | standard |
| 3.84704 | 1.0 | standard |
| 3.88759 | 0.999397 | standard |
| 6.64128 | 0.181504 | standard |
| 6.65542 | 0.181854 | standard |
| 8.21755 | 0.181936 | standard |
| 8.23159 | 0.181783 | standard |
| 3.84704 | 1.0 | standard |
| 3.8876 | 0.999915 | standard |
| 6.64371 | 0.182913 | standard |
| 6.6531 | 0.18341 | standard |
| 8.21988 | 0.183354 | standard |
| 8.22927 | 0.182985 | standard |
| 3.84703 | 1.0 | standard |
| 3.8876 | 0.998699 | standard |
| 6.64487 | 0.183641 | standard |
| 6.65194 | 0.183395 | standard |
| 8.22104 | 0.183524 | standard |
| 8.22811 | 0.182985 | standard |
| 3.84703 | 0.999467 | standard |
| 3.8876 | 1.0 | standard |
| 6.64555 | 0.18406 | standard |
| 6.65116 | 0.184215 | standard |
| 8.22172 | 0.184277 | standard |
| 8.22733 | 0.184379 | standard |
| 3.84703 | 1.0 | standard |
| 3.8876 | 0.999501 | standard |
| 6.64603 | 0.183851 | standard |
| 6.65078 | 0.183381 | standard |
| 8.2222 | 0.183997 | standard |
| 8.22684 | 0.184272 | standard |
| 3.84703 | 0.999873 | standard |
| 3.8876 | 1.0 | standard |
| 6.64632 | 0.183787 | standard |
| 6.65039 | 0.183925 | standard |
| 8.22249 | 0.18369 | standard |
| 8.22656 | 0.183747 | standard |
| 3.84703 | 1.0 | standard |
| 3.8876 | 0.999612 | standard |
| 6.64651 | 0.184034 | standard |
| 6.65029 | 0.184219 | standard |
| 8.22268 | 0.184024 | standard |
| 8.22646 | 0.184349 | standard |
| 3.84703 | 0.99986 | standard |
| 3.8876 | 1.0 | standard |
| 6.64661 | 0.184479 | standard |
| 6.65009 | 0.184111 | standard |
| 8.22278 | 0.184941 | standard |
| 8.22626 | 0.184307 | standard |
| 3.84703 | 1.0 | standard |
| 3.8876 | 0.999732 | standard |
| 6.64681 | 0.184848 | standard |
| 6.6499 | 0.184631 | standard |
| 8.22298 | 0.18492 | standard |
| 8.22607 | 0.184714 | standard |
| 3.84703 | 1.0 | standard |
| 3.8876 | 0.998991 | standard |
| 6.6469 | 0.183753 | standard |
| 6.6499 | 0.183487 | standard |
| 8.22307 | 0.184686 | standard |
| 8.22597 | 0.184563 | standard |
| 3.84703 | 1.0 | standard |
| 3.8876 | 0.99955 | standard |
| 6.647 | 0.184657 | standard |
| 6.64981 | 0.184341 | standard |
| 8.22317 | 0.184434 | standard |
| 8.22598 | 0.184211 | standard |
| 3.84703 | 1.0 | standard |
| 3.8876 | 0.99986 | standard |
| 6.64729 | 0.184674 | standard |
| 6.64942 | 0.184928 | standard |
| 8.22346 | 0.184647 | standard |
| 8.22559 | 0.184699 | standard |