N1-[3-(ethylamino)-4-methylphenyl]acetamide
Simulation outputs:
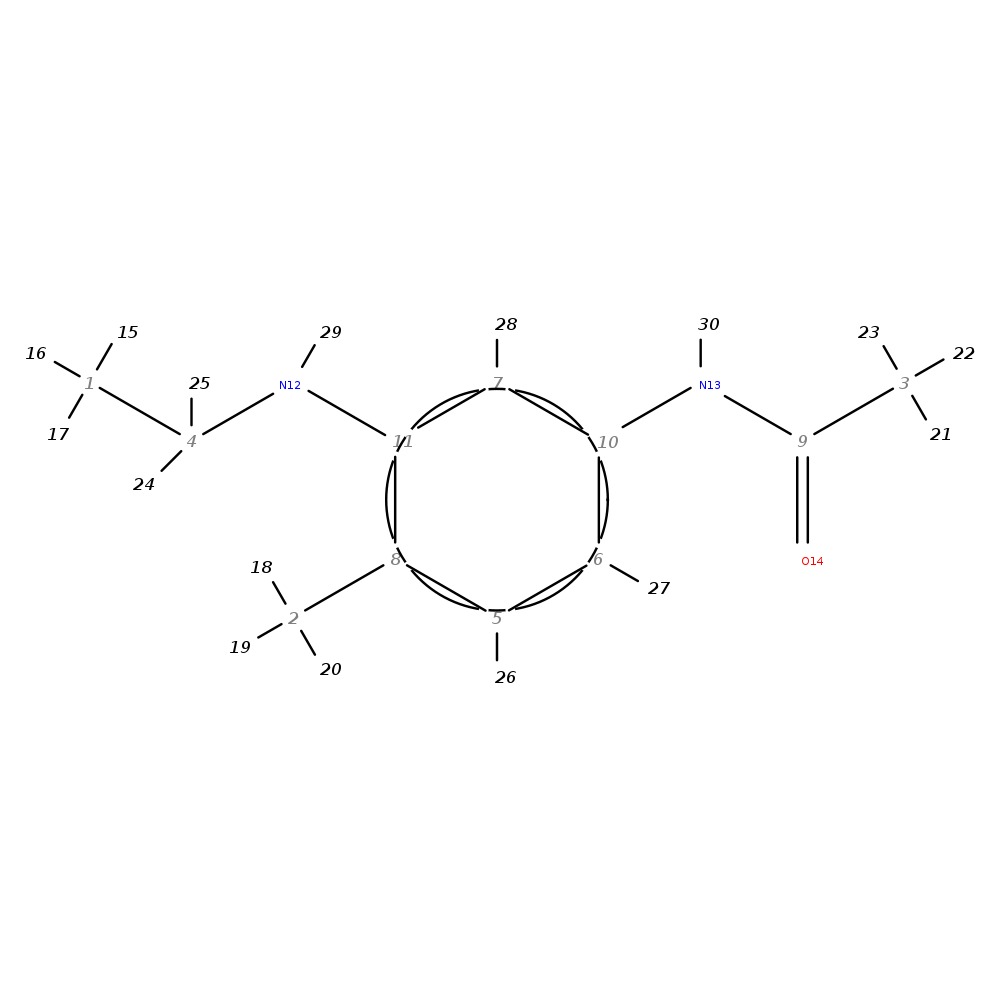
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01785 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H16N2O/c1-4-12-11-7-10(13-9(3)14)6-5-8(11)2/h5-7,12H,4H2,1-3H3,(H,13,14) | |
| Note 1 | 18,19,20?21,22,23 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N1-[3-(ethylamino)-4-methylphenyl]acetamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 1.229 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.445 | 7.445 | 0 | 0 | 0 |
| 16 | 0 | 1.229 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.445 | 7.445 | 0 | 0 | 0 |
| 17 | 0 | 0 | 1.229 | 0 | 0 | 0 | 0 | 0 | 0 | 7.445 | 7.445 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 2.129 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 2.129 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 2.129 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 2.144 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.144 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.144 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.141 | -14.0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.141 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.123 | 8.445 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.768 | 1.438 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.872 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.21645 | 0.256938 | standard |
| 1.22886 | 0.50231 | standard |
| 1.24128 | 0.256997 | standard |
| 2.12873 | 0.99997 | standard |
| 2.1437 | 1.0 | standard |
| 3.12244 | 0.0880337 | standard |
| 3.13483 | 0.252711 | standard |
| 3.14725 | 0.25298 | standard |
| 3.15961 | 0.0880616 | standard |
| 6.76042 | 0.110775 | standard |
| 6.76207 | 0.111661 | standard |
| 6.77448 | 0.117757 | standard |
| 6.77613 | 0.117666 | standard |
| 6.87129 | 0.22398 | standard |
| 6.87294 | 0.223498 | standard |
| 7.11632 | 0.171619 | standard |
| 7.1304 | 0.162071 | standard |
| 1.03694 | 0.0978581 | standard |
| 1.22137 | 0.243845 | standard |
| 1.40383 | 0.147905 | standard |
| 2.13622 | 1.0 | standard |
| 2.87754 | 0.0536161 | standard |
| 3.0563 | 0.124388 | standard |
| 3.24228 | 0.105845 | standard |
| 3.42673 | 0.0299151 | standard |
| 6.64612 | 0.0380841 | standard |
| 6.85875 | 0.268287 | standard |
| 7.04912 | 0.131243 | standard |
| 7.25473 | 0.0438093 | standard |
| 1.10162 | 0.12549 | standard |
| 1.22506 | 0.288672 | standard |
| 1.34832 | 0.163312 | standard |
| 2.13621 | 1.0 | standard |
| 2.9617 | 0.058528 | standard |
| 3.08404 | 0.146174 | standard |
| 3.20761 | 0.130618 | standard |
| 3.33211 | 0.0404171 | standard |
| 6.69319 | 0.050203 | standard |
| 6.83686 | 0.152414 | standard |
| 6.86259 | 0.185515 | standard |
| 7.06732 | 0.136517 | standard |
| 7.20595 | 0.0645731 | standard |
| 1.13392 | 0.152043 | standard |
| 1.22668 | 0.334051 | standard |
| 1.31947 | 0.184221 | standard |
| 2.13381 | 0.999893 | standard |
| 2.13867 | 1.0 | standard |
| 3.00515 | 0.065559 | standard |
| 3.09767 | 0.170031 | standard |
| 3.19041 | 0.155814 | standard |
| 3.28354 | 0.049644 | standard |
| 6.71451 | 0.061765 | standard |
| 6.82024 | 0.125437 | standard |
| 6.86556 | 0.179347 | standard |
| 7.07853 | 0.147522 | standard |
| 7.18323 | 0.0829458 | standard |
| 1.14465 | 0.164028 | standard |
| 1.22713 | 0.354795 | standard |
| 1.30965 | 0.193785 | standard |
| 2.13161 | 1.0 | standard |
| 2.14081 | 0.9995 | standard |
| 3.01987 | 0.068849 | standard |
| 3.10216 | 0.180931 | standard |
| 3.1847 | 0.167503 | standard |
| 3.26744 | 0.0539649 | standard |
| 6.72117 | 0.067334 | standard |
| 6.81502 | 0.122831 | standard |
| 6.86639 | 0.183658 | standard |
| 7.0827 | 0.152248 | standard |
| 7.17594 | 0.0915541 | standard |
| 1.1532 | 0.174126 | standard |
| 1.22747 | 0.37281 | standard |
| 1.30178 | 0.202274 | standard |
| 2.13064 | 1.0 | standard |
| 2.14178 | 0.999073 | standard |
| 3.03171 | 0.071789 | standard |
| 3.1059 | 0.190106 | standard |
| 3.18015 | 0.177046 | standard |
| 3.25457 | 0.0575032 | standard |
| 6.72647 | 0.0719776 | standard |
| 6.81088 | 0.121603 | standard |
| 6.86694 | 0.187858 | standard |
| 7.08617 | 0.156259 | standard |
| 7.17014 | 0.0989768 | standard |
| 1.19135 | 0.228582 | standard |
| 1.22855 | 0.459049 | standard |
| 1.26572 | 0.240501 | standard |
| 2.12888 | 1.00007 | standard |
| 2.14355 | 0.998719 | standard |
| 3.08577 | 0.0841811 | standard |
| 3.12293 | 0.232096 | standard |
| 3.16011 | 0.227514 | standard |
| 3.19725 | 0.0777593 | standard |
| 6.74383 | 0.091183 | standard |
| 6.74866 | 0.0959881 | standard |
| 6.78608 | 0.114443 | standard |
| 6.79082 | 0.121608 | standard |
| 6.86957 | 0.212839 | standard |
| 6.87433 | 0.201013 | standard |
| 7.10338 | 0.170882 | standard |
| 7.14556 | 0.13565 | standard |
| 1.20394 | 0.245718 | standard |
| 1.22876 | 0.484494 | standard |
| 1.25353 | 0.248623 | standard |
| 2.12876 | 1.0001 | standard |
| 2.14367 | 0.998386 | standard |
| 3.10401 | 0.0846453 | standard |
| 3.12881 | 0.242027 | standard |
| 3.15362 | 0.244692 | standard |
| 3.17844 | 0.0846653 | standard |
| 6.75211 | 0.0997701 | standard |
| 6.75557 | 0.103093 | standard |
| 6.78034 | 0.117176 | standard |
| 6.78361 | 0.121403 | standard |
| 6.87041 | 0.219453 | standard |
| 6.87377 | 0.212317 | standard |
| 7.10972 | 0.173767 | standard |
| 7.13783 | 0.149254 | standard |
| 1.2102 | 0.252634 | standard |
| 1.22881 | 0.494738 | standard |
| 1.24742 | 0.252882 | standard |
| 2.12874 | 1.00002 | standard |
| 2.1437 | 0.999028 | standard |
| 3.11322 | 0.0864867 | standard |
| 3.13184 | 0.247564 | standard |
| 3.15039 | 0.249773 | standard |
| 3.16902 | 0.0865116 | standard |
| 6.75637 | 0.10576 | standard |
| 6.75884 | 0.107488 | standard |
| 6.77734 | 0.11672 | standard |
| 6.77999 | 0.118768 | standard |
| 6.87083 | 0.221444 | standard |
| 6.87338 | 0.21795 | standard |
| 7.113 | 0.173497 | standard |
| 7.13406 | 0.155243 | standard |
| 1.21394 | 0.255202 | standard |
| 1.22886 | 0.499754 | standard |
| 1.2437 | 0.254972 | standard |
| 2.12873 | 1.00001 | standard |
| 2.1437 | 0.999016 | standard |
| 3.11872 | 0.0873573 | standard |
| 3.13364 | 0.25139 | standard |
| 3.14851 | 0.251483 | standard |
| 3.16343 | 0.0873583 | standard |
| 6.7588 | 0.108945 | standard |
| 6.76082 | 0.109955 | standard |
| 6.77565 | 0.117244 | standard |
| 6.77767 | 0.118063 | standard |
| 6.87115 | 0.222842 | standard |
| 6.87316 | 0.221411 | standard |
| 7.11503 | 0.172049 | standard |
| 7.13188 | 0.159836 | standard |
| 1.21645 | 0.256449 | standard |
| 1.22886 | 0.50231 | standard |
| 1.24128 | 0.256873 | standard |
| 2.12873 | 1.0 | standard |
| 2.1437 | 0.998945 | standard |
| 3.12244 | 0.0879927 | standard |
| 3.13483 | 0.252713 | standard |
| 3.14725 | 0.252854 | standard |
| 3.15961 | 0.0880186 | standard |
| 6.76042 | 0.110615 | standard |
| 6.76207 | 0.111409 | standard |
| 6.77448 | 0.117572 | standard |
| 6.77613 | 0.117401 | standard |
| 6.87129 | 0.224293 | standard |
| 6.87293 | 0.223462 | standard |
| 7.11632 | 0.171539 | standard |
| 7.1304 | 0.162022 | standard |
| 1.21823 | 0.257372 | standard |
| 1.22886 | 0.503644 | standard |
| 1.23949 | 0.257625 | standard |
| 2.12873 | 0.999402 | standard |
| 2.14371 | 1.0 | standard |
| 3.12512 | 0.0882915 | standard |
| 3.1357 | 0.2539 | standard |
| 3.14631 | 0.253784 | standard |
| 3.15693 | 0.0882588 | standard |
| 6.76154 | 0.111825 | standard |
| 6.76302 | 0.112455 | standard |
| 6.77362 | 0.115622 | standard |
| 6.77511 | 0.115392 | standard |
| 6.87142 | 0.22258 | standard |
| 6.87291 | 0.222368 | standard |
| 7.11731 | 0.169771 | standard |
| 7.1294 | 0.164989 | standard |
| 1.21897 | 0.257282 | standard |
| 1.22886 | 0.503961 | standard |
| 1.2388 | 0.25766 | standard |
| 2.12873 | 1.0 | standard |
| 2.14371 | 0.998559 | standard |
| 3.12611 | 0.0882084 | standard |
| 3.13608 | 0.254014 | standard |
| 3.146 | 0.254085 | standard |
| 3.15589 | 0.0884054 | standard |
| 6.76199 | 0.111144 | standard |
| 6.76345 | 0.1117 | standard |
| 6.77333 | 0.117248 | standard |
| 6.7746 | 0.116713 | standard |
| 6.87153 | 0.225738 | standard |
| 6.8728 | 0.22588 | standard |
| 7.11771 | 0.169786 | standard |
| 7.12891 | 0.165317 | standard |
| 1.21957 | 0.257863 | standard |
| 1.22886 | 0.50437 | standard |
| 1.2382 | 0.257893 | standard |
| 2.12873 | 1.0 | standard |
| 2.14371 | 0.998277 | standard |
| 3.1271 | 0.0883776 | standard |
| 3.13638 | 0.254388 | standard |
| 3.14565 | 0.254085 | standard |
| 3.15495 | 0.0883576 | standard |
| 6.76236 | 0.11062 | standard |
| 6.76378 | 0.111385 | standard |
| 6.773 | 0.116418 | standard |
| 6.77423 | 0.115957 | standard |
| 6.8715 | 0.22513 | standard |
| 6.87273 | 0.224998 | standard |
| 7.118 | 0.169311 | standard |
| 7.12861 | 0.165362 | standard |
| 1.22061 | 0.257927 | standard |
| 1.22891 | 0.506075 | standard |
| 1.23716 | 0.258314 | standard |
| 2.12873 | 0.998709 | standard |
| 2.14371 | 1.0 | standard |
| 3.12859 | 0.0884054 | standard |
| 3.13688 | 0.254537 | standard |
| 3.14512 | 0.25439 | standard |
| 3.15341 | 0.0885733 | standard |
| 6.76317 | 0.114219 | standard |
| 6.76426 | 0.114765 | standard |
| 6.77247 | 0.116137 | standard |
| 6.77357 | 0.115538 | standard |
| 6.87157 | 0.225157 | standard |
| 6.87266 | 0.225014 | standard |
| 7.1186 | 0.168712 | standard |
| 7.12802 | 0.167008 | standard |
| 1.22106 | 0.258337 | standard |
| 1.22891 | 0.506308 | standard |
| 1.23672 | 0.25855 | standard |
| 2.12873 | 0.999757 | standard |
| 2.14371 | 1.0 | standard |
| 3.12928 | 0.0885248 | standard |
| 3.13709 | 0.254677 | standard |
| 3.14492 | 0.254326 | standard |
| 3.15277 | 0.0884795 | standard |
| 6.76346 | 0.114664 | standard |
| 6.76447 | 0.115377 | standard |
| 6.77227 | 0.116698 | standard |
| 6.77328 | 0.116155 | standard |
| 6.87166 | 0.226719 | standard |
| 6.87267 | 0.226734 | standard |
| 7.1188 | 0.168998 | standard |
| 7.12772 | 0.167094 | standard |
| 1.22145 | 0.25806 | standard |
| 1.22891 | 0.506129 | standard |
| 1.23634 | 0.258747 | standard |
| 2.12873 | 1.0 | standard |
| 2.14371 | 0.998085 | standard |
| 3.12983 | 0.0886163 | standard |
| 3.13727 | 0.254689 | standard |
| 3.14473 | 0.254481 | standard |
| 3.15218 | 0.0884096 | standard |
| 6.76362 | 0.112622 | standard |
| 6.7647 | 0.113232 | standard |
| 6.77203 | 0.113763 | standard |
| 6.77312 | 0.113213 | standard |
| 6.87166 | 0.228401 | standard |
| 6.87257 | 0.228364 | standard |
| 7.11909 | 0.168618 | standard |
| 7.12752 | 0.167515 | standard |
| 1.22316 | 0.259335 | standard |
| 1.22891 | 0.507653 | standard |
| 1.23464 | 0.258685 | standard |
| 2.12873 | 1.0 | standard |
| 2.14371 | 0.999755 | standard |
| 3.1324 | 0.0888373 | standard |
| 3.13815 | 0.255467 | standard |
| 3.14387 | 0.254711 | standard |
| 3.14955 | 0.0889536 | standard |
| 6.76478 | 0.115421 | standard |
| 6.76552 | 0.115813 | standard |
| 6.77118 | 0.112207 | standard |
| 6.7721 | 0.1115 | standard |
| 6.8718 | 0.226587 | standard |
| 6.87253 | 0.226537 | standard |
| 7.11999 | 0.168607 | standard |
| 7.12653 | 0.1685 | standard |