Methyl 3-amino-4-methylbenzoate
Simulation outputs:
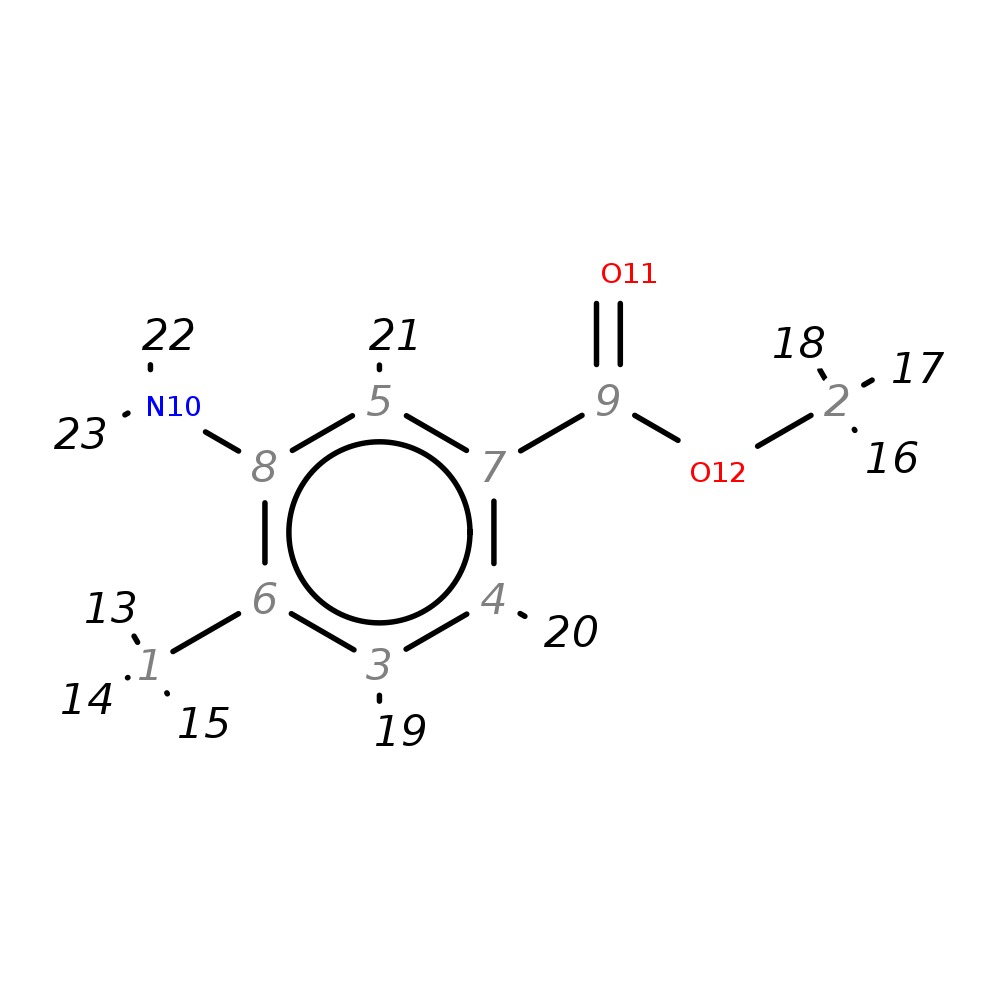
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02364 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H11NO2/c1-6-3-4-7(5-8(6)10)9(11)12-2/h3-5H,10H2,1-2H3 | |
| Note 1 | 13,14,15?16,17,18 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Methyl 3-amino-4-methylbenzoate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|
| 13 | 2.224 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.224 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.224 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 3.89 | -14.0 | -14.0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 3.89 | -14.0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 3.89 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 7.246 | 8.756 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.424 | 1.2 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.428 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.999246 | standard |
| 7.23868 | 0.146484 | standard |
| 7.25362 | 0.174145 | standard |
| 7.41777 | 0.150376 | standard |
| 7.42847 | 0.297593 | standard |
| 2.22435 | 0.997954 | standard |
| 3.88968 | 1.0 | standard |
| 7.08319 | 0.043158 | standard |
| 7.30728 | 0.350952 | standard |
| 7.37561 | 0.351438 | standard |
| 7.4164 | 0.354466 | standard |
| 7.57649 | 0.0468661 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.998763 | standard |
| 7.14617 | 0.06434 | standard |
| 7.29466 | 0.284008 | standard |
| 7.38365 | 0.277257 | standard |
| 7.42142 | 0.310852 | standard |
| 7.51668 | 0.0669242 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.999716 | standard |
| 7.17528 | 0.0802191 | standard |
| 7.28663 | 0.25669 | standard |
| 7.38986 | 0.244396 | standard |
| 7.42344 | 0.292941 | standard |
| 7.48921 | 0.0829065 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.997919 | standard |
| 7.18454 | 0.0869731 | standard |
| 7.28349 | 0.24697 | standard |
| 7.39236 | 0.233244 | standard |
| 7.424 | 0.287176 | standard |
| 7.48054 | 0.0893501 | standard |
| 2.22435 | 0.998413 | standard |
| 3.88968 | 1.0 | standard |
| 7.19172 | 0.0932634 | standard |
| 7.28076 | 0.238307 | standard |
| 7.39458 | 0.224586 | standard |
| 7.42449 | 0.283469 | standard |
| 7.4739 | 0.0954021 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.998922 | standard |
| 7.22153 | 0.124612 | standard |
| 7.24438 | 0.0268351 | standard |
| 7.26602 | 0.201852 | standard |
| 7.40697 | 0.182966 | standard |
| 7.42686 | 0.270334 | standard |
| 7.44639 | 0.129478 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.999629 | standard |
| 7.2304 | 0.135564 | standard |
| 7.24322 | 0.0234194 | standard |
| 7.26007 | 0.18922 | standard |
| 7.41206 | 0.16853 | standard |
| 7.4285 | 0.269602 | standard |
| 7.43823 | 0.149283 | standard |
| 2.22435 | 0.999573 | standard |
| 3.88968 | 1.0 | standard |
| 7.23466 | 0.14191 | standard |
| 7.25695 | 0.18191 | standard |
| 7.41491 | 0.160344 | standard |
| 7.42858 | 0.274351 | standard |
| 7.43438 | 0.167518 | standard |
| 2.22435 | 0.99989 | standard |
| 3.88968 | 1.0 | standard |
| 7.23709 | 0.145453 | standard |
| 7.25496 | 0.176647 | standard |
| 7.41664 | 0.155003 | standard |
| 7.4285 | 0.283772 | standard |
| 7.43183 | 0.190794 | standard |
| 2.22435 | 0.998766 | standard |
| 3.88968 | 1.0 | standard |
| 7.23868 | 0.146449 | standard |
| 7.25362 | 0.174104 | standard |
| 7.41777 | 0.150341 | standard |
| 7.42847 | 0.297523 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.998893 | standard |
| 7.23987 | 0.147051 | standard |
| 7.25263 | 0.170561 | standard |
| 7.41869 | 0.147929 | standard |
| 7.42847 | 0.315713 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.999931 | standard |
| 7.24027 | 0.147 | standard |
| 7.25223 | 0.16898 | standard |
| 7.419 | 0.146247 | standard |
| 7.42852 | 0.334385 | standard |
| 2.22435 | 0.999492 | standard |
| 3.88968 | 1.0 | standard |
| 7.24067 | 0.146755 | standard |
| 7.25187 | 0.165457 | standard |
| 7.4193 | 0.14515 | standard |
| 7.42845 | 0.346292 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.99877 | standard |
| 7.24134 | 0.144075 | standard |
| 7.25132 | 0.162638 | standard |
| 7.41981 | 0.144841 | standard |
| 7.42833 | 0.387629 | standard |
| 2.22435 | 1.0 | standard |
| 3.88968 | 0.999911 | standard |
| 7.24159 | 0.145232 | standard |
| 7.25105 | 0.158657 | standard |
| 7.42 | 0.145212 | standard |
| 7.42829 | 0.403028 | standard |
| 2.22435 | 0.999903 | standard |
| 3.88968 | 1.0 | standard |
| 7.24186 | 0.142673 | standard |
| 7.25084 | 0.156561 | standard |
| 7.42023 | 0.142147 | standard |
| 7.42816 | 0.418791 | standard |
| 2.22435 | 0.999304 | standard |
| 3.88968 | 1.0 | standard |
| 7.24306 | 0.142174 | standard |
| 7.24966 | 0.157183 | standard |
| 7.42114 | 0.139985 | standard |
| 7.42765 | 0.455002 | standard |