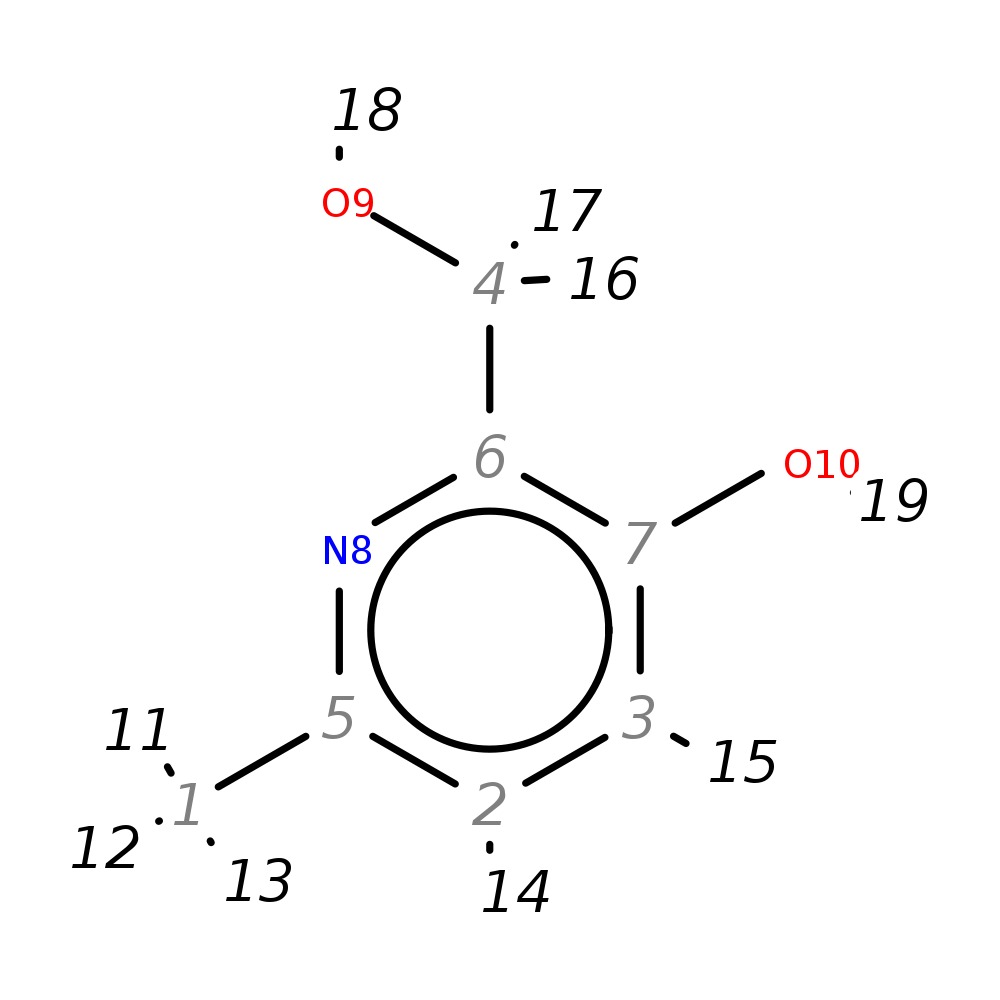
2-(Hydroxymethyl)-6-methylpyridin-3-ol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.15203 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H9NO2/c1-5-2-3-7(10)6(4-9)8-5/h2-3,9-10H,4H2,1H3 | |
| Note 1 | 16,17 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(Hydroxymethyl)-6-methylpyridin-3-ol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|
| 11 | 2.482 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 2.482 | -14.0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 2.482 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 7.248 | 8.065 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 7.323 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 4.36 | 11.89 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 4.354 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.48183 | 1.0 | standard |
| 4.33701 | 0.0111181 | standard |
| 4.3572 | 0.608802 | standard |
| 4.3774 | 0.0115625 | standard |
| 7.2409 | 0.140432 | standard |
| 7.25438 | 0.198259 | standard |
| 7.31727 | 0.19826 | standard |
| 7.33075 | 0.140431 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.666554 | standard |
| 7.08036 | 0.0198561 | standard |
| 7.28582 | 0.602078 | standard |
| 7.49159 | 0.0198391 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.666549 | standard |
| 7.1424 | 0.029673 | standard |
| 7.28344 | 0.464039 | standard |
| 7.28825 | 0.464039 | standard |
| 7.4293 | 0.0296672 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.666412 | standard |
| 7.17288 | 0.0406851 | standard |
| 7.27445 | 0.36328 | standard |
| 7.2972 | 0.36328 | standard |
| 7.39877 | 0.0406831 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.666331 | standard |
| 7.1828 | 0.0462243 | standard |
| 7.27271 | 0.337651 | standard |
| 7.29893 | 0.33765 | standard |
| 7.38885 | 0.0462223 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.666242 | standard |
| 7.19058 | 0.0516343 | standard |
| 7.27136 | 0.319524 | standard |
| 7.30028 | 0.319525 | standard |
| 7.38107 | 0.0516333 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.664398 | standard |
| 7.22307 | 0.092615 | standard |
| 7.26331 | 0.250051 | standard |
| 7.30834 | 0.250051 | standard |
| 7.34858 | 0.092615 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.65924 | standard |
| 7.23247 | 0.114427 | standard |
| 7.25934 | 0.225514 | standard |
| 7.31231 | 0.225514 | standard |
| 7.33917 | 0.114427 | standard |
| 2.48183 | 1.0 | standard |
| 4.35725 | 0.650858 | standard |
| 7.23683 | 0.12699 | standard |
| 7.25696 | 0.212222 | standard |
| 7.31469 | 0.212222 | standard |
| 7.33481 | 0.12699 | standard |
| 2.48183 | 1.0 | standard |
| 4.3572 | 0.637335 | standard |
| 7.23931 | 0.134951 | standard |
| 7.25547 | 0.203912 | standard |
| 7.31618 | 0.203912 | standard |
| 7.33234 | 0.134951 | standard |
| 2.48183 | 1.0 | standard |
| 4.337 | 0.0115634 | standard |
| 4.3572 | 0.608805 | standard |
| 4.3774 | 0.0115634 | standard |
| 7.2409 | 0.140425 | standard |
| 7.25438 | 0.198259 | standard |
| 7.31727 | 0.198252 | standard |
| 7.33075 | 0.14043 | standard |
| 2.48183 | 1.0 | standard |
| 4.33975 | 0.0138136 | standard |
| 4.35725 | 0.578053 | standard |
| 4.37465 | 0.0138642 | standard |
| 7.24199 | 0.144372 | standard |
| 7.25359 | 0.194161 | standard |
| 7.31816 | 0.194196 | standard |
| 7.32966 | 0.144421 | standard |
| 2.48183 | 1.0 | standard |
| 4.34083 | 0.015038 | standard |
| 4.3572 | 0.552649 | standard |
| 4.37357 | 0.0150383 | standard |
| 7.24248 | 0.14575 | standard |
| 7.25319 | 0.192559 | standard |
| 7.31846 | 0.192145 | standard |
| 7.32916 | 0.146059 | standard |
| 2.48183 | 1.0 | standard |
| 4.34181 | 0.0163325 | standard |
| 4.35725 | 0.523881 | standard |
| 4.37259 | 0.0163868 | standard |
| 7.24288 | 0.1475 | standard |
| 7.25289 | 0.191134 | standard |
| 7.31876 | 0.191134 | standard |
| 7.32877 | 0.1475 | standard |
| 2.48183 | 1.0 | standard |
| 4.34339 | 0.0190554 | standard |
| 4.3572 | 0.476632 | standard |
| 4.37101 | 0.0190554 | standard |
| 7.24347 | 0.149856 | standard |
| 7.2524 | 0.188712 | standard |
| 7.31925 | 0.188712 | standard |
| 7.32817 | 0.149856 | standard |
| 2.48183 | 1.0 | standard |
| 4.34408 | 0.0204125 | standard |
| 4.35686 | 0.446514 | standard |
| 4.35764 | 0.446512 | standard |
| 4.37042 | 0.0204535 | standard |
| 7.24377 | 0.150503 | standard |
| 7.2522 | 0.187701 | standard |
| 7.31945 | 0.187701 | standard |
| 7.32788 | 0.150503 | standard |
| 2.48183 | 1.0 | standard |
| 4.34467 | 0.0217838 | standard |
| 4.35679 | 0.4346 | standard |
| 4.3577 | 0.434599 | standard |
| 4.36983 | 0.0218928 | standard |
| 7.24397 | 0.151778 | standard |
| 7.252 | 0.186777 | standard |
| 7.31965 | 0.186777 | standard |
| 7.32768 | 0.151778 | standard |
| 2.48183 | 1.0 | standard |
| 4.34714 | 0.0309579 | standard |
| 4.3564 | 0.355325 | standard |
| 4.35809 | 0.355327 | standard |
| 4.36726 | 0.0310311 | standard |
| 7.24496 | 0.155813 | standard |
| 7.25121 | 0.18256 | standard |
| 7.32044 | 0.18256 | standard |
| 7.32669 | 0.155813 | standard |