2-(Methylthio)nicotinic acid
Simulation outputs:
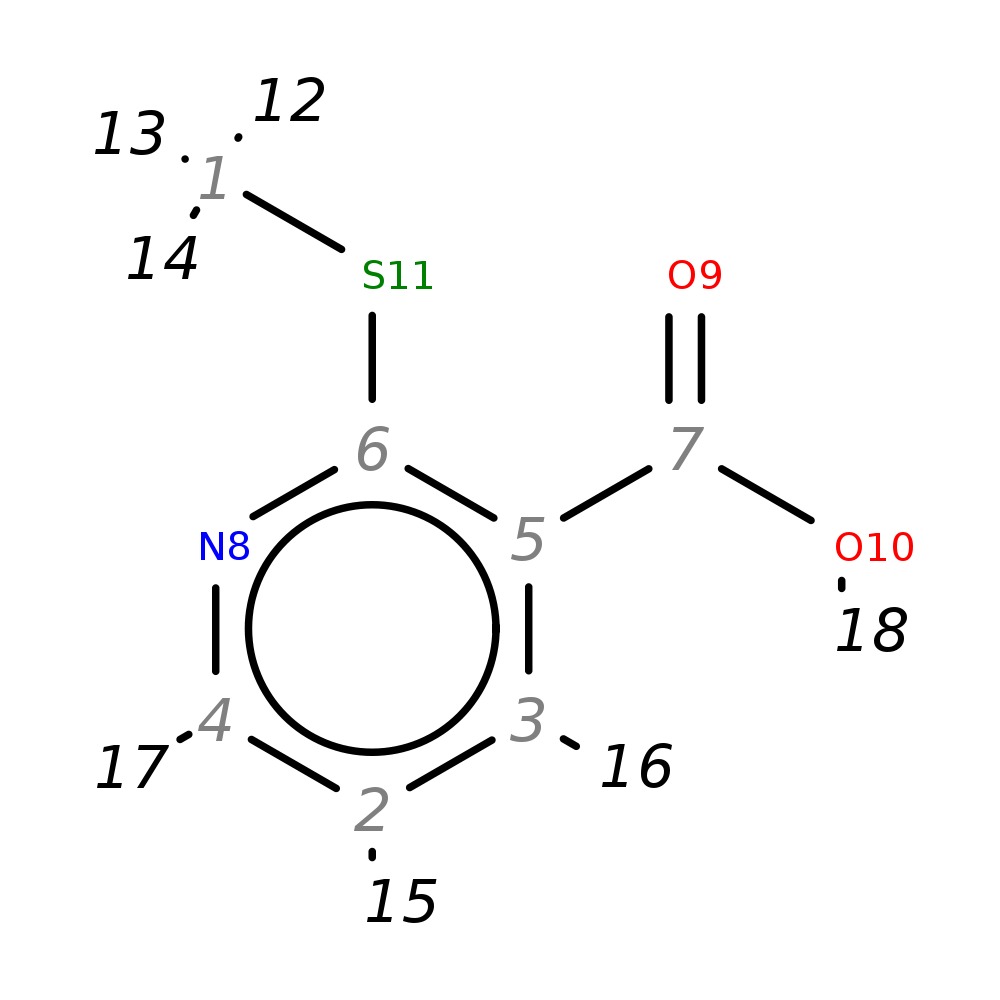
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.04470 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H7NO2S/c1-11-6-5(7(9)10)3-2-4-8-6/h2-4H,1H3,(H,9,10) | |
| Note 1 | 16?17 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(Methylthio)nicotinic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|
| 12 | 2.486 | -14.0 | -14.0 | 0 | 0 | 0 |
| 13 | 0 | 2.486 | -14.0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 2.486 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.208 | 8.124 | 5.26 |
| 16 | 0 | 0 | 0 | 0 | 7.852 | 1.51 |
| 17 | 0 | 0 | 0 | 0 | 0 | 8.41 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.4858 | 1.0 | standard |
| 7.19657 | 0.0874631 | standard |
| 7.20544 | 0.0961111 | standard |
| 7.21009 | 0.0969276 | standard |
| 7.21886 | 0.0884324 | standard |
| 7.84455 | 0.112379 | standard |
| 7.84654 | 0.113157 | standard |
| 7.85815 | 0.113858 | standard |
| 7.85997 | 0.11315 | standard |
| 8.40491 | 0.116413 | standard |
| 8.40675 | 0.119131 | standard |
| 8.41355 | 0.119285 | standard |
| 8.41539 | 0.116556 | standard |
| 2.4858 | 1.0 | standard |
| 7.02323 | 0.0582154 | standard |
| 7.15217 | 0.0766142 | standard |
| 7.22592 | 0.107608 | standard |
| 7.35411 | 0.127434 | standard |
| 7.75026 | 0.135315 | standard |
| 7.78323 | 0.151098 | standard |
| 7.95181 | 0.0801161 | standard |
| 7.98538 | 0.078898 | standard |
| 8.33318 | 0.129351 | standard |
| 8.36691 | 0.122731 | standard |
| 8.46021 | 0.108331 | standard |
| 8.49401 | 0.098925 | standard |
| 2.4858 | 1.0 | standard |
| 7.08832 | 0.0668011 | standard |
| 7.17542 | 0.083938 | standard |
| 7.22334 | 0.104919 | standard |
| 7.31011 | 0.113587 | standard |
| 7.78168 | 0.129842 | standard |
| 7.80209 | 0.138666 | standard |
| 7.9164 | 0.0901744 | standard |
| 7.9369 | 0.090153 | standard |
| 8.3583 | 0.124913 | standard |
| 8.37911 | 0.122132 | standard |
| 8.4442 | 0.111759 | standard |
| 8.46493 | 0.104929 | standard |
| 2.4858 | 1.0 | standard |
| 7.11962 | 0.0715903 | standard |
| 7.18534 | 0.0874661 | standard |
| 7.22079 | 0.103129 | standard |
| 7.28633 | 0.106861 | standard |
| 7.79824 | 0.126628 | standard |
| 7.81297 | 0.132405 | standard |
| 7.89934 | 0.0960866 | standard |
| 7.91419 | 0.0957261 | standard |
| 8.37113 | 0.122702 | standard |
| 8.38612 | 0.121773 | standard |
| 8.43584 | 0.113184 | standard |
| 8.45096 | 0.108093 | standard |
| 2.4858 | 1.0 | standard |
| 7.12991 | 0.0735531 | standard |
| 7.18835 | 0.0883153 | standard |
| 7.21982 | 0.102644 | standard |
| 7.2781 | 0.103928 | standard |
| 7.80384 | 0.125093 | standard |
| 7.8169 | 0.129551 | standard |
| 7.89372 | 0.0980862 | standard |
| 7.90683 | 0.096991 | standard |
| 8.37542 | 0.12094 | standard |
| 8.38868 | 0.121216 | standard |
| 8.43304 | 0.11386 | standard |
| 8.44625 | 0.109062 | standard |
| 2.4858 | 1.0 | standard |
| 7.13804 | 0.074887 | standard |
| 7.19068 | 0.0892223 | standard |
| 7.21894 | 0.102245 | standard |
| 7.27146 | 0.102597 | standard |
| 7.80833 | 0.124282 | standard |
| 7.82013 | 0.127951 | standard |
| 7.88941 | 0.100123 | standard |
| 7.90108 | 0.099274 | standard |
| 8.37884 | 0.120634 | standard |
| 8.39064 | 0.121312 | standard |
| 8.43078 | 0.114806 | standard |
| 8.44255 | 0.11098 | standard |
| 2.4858 | 1.0 | standard |
| 7.17369 | 0.0823523 | standard |
| 7.2 | 0.0920969 | standard |
| 7.21408 | 0.100819 | standard |
| 7.24045 | 0.093856 | standard |
| 7.82974 | 0.119946 | standard |
| 7.8354 | 0.120738 | standard |
| 7.8702 | 0.106638 | standard |
| 7.87603 | 0.10573 | standard |
| 8.39434 | 0.115932 | standard |
| 8.40023 | 0.118827 | standard |
| 8.42037 | 0.117709 | standard |
| 8.42626 | 0.114778 | standard |
| 2.4858 | 1.0 | standard |
| 7.18518 | 0.0846782 | standard |
| 7.20282 | 0.0936252 | standard |
| 7.21213 | 0.0995123 | standard |
| 7.22976 | 0.0912402 | standard |
| 7.83704 | 0.116956 | standard |
| 7.84087 | 0.117655 | standard |
| 7.86408 | 0.109398 | standard |
| 7.86791 | 0.108624 | standard |
| 8.39957 | 0.115527 | standard |
| 8.40344 | 0.118402 | standard |
| 8.41696 | 0.118377 | standard |
| 8.42083 | 0.115581 | standard |
| 2.4858 | 1.0 | standard |
| 7.19092 | 0.0861891 | standard |
| 7.20418 | 0.09506 | standard |
| 7.21116 | 0.0982661 | standard |
| 7.22431 | 0.0897954 | standard |
| 7.84086 | 0.115421 | standard |
| 7.84369 | 0.11615 | standard |
| 7.86103 | 0.110538 | standard |
| 7.86402 | 0.109732 | standard |
| 8.40215 | 0.11475 | standard |
| 8.40518 | 0.117713 | standard |
| 8.41525 | 0.118759 | standard |
| 8.41811 | 0.115949 | standard |
| 2.4858 | 1.0 | standard |
| 7.19429 | 0.087025 | standard |
| 7.20495 | 0.096 | standard |
| 7.21048 | 0.0976229 | standard |
| 7.22104 | 0.0888703 | standard |
| 7.84302 | 0.113383 | standard |
| 7.84539 | 0.11414 | standard |
| 7.85927 | 0.111896 | standard |
| 7.86163 | 0.111123 | standard |
| 8.40375 | 0.115032 | standard |
| 8.40614 | 0.117968 | standard |
| 8.41417 | 0.11797 | standard |
| 8.41655 | 0.115038 | standard |
| 2.4858 | 1.0 | standard |
| 7.19657 | 0.087469 | standard |
| 7.20544 | 0.0962321 | standard |
| 7.21009 | 0.0970708 | standard |
| 7.21886 | 0.0884384 | standard |
| 7.84455 | 0.112544 | standard |
| 7.84654 | 0.113306 | standard |
| 7.85815 | 0.113863 | standard |
| 7.85997 | 0.113154 | standard |
| 8.40491 | 0.116564 | standard |
| 8.40675 | 0.119298 | standard |
| 8.41355 | 0.119298 | standard |
| 8.41539 | 0.116564 | standard |
| 2.4858 | 1.0 | standard |
| 7.19825 | 0.0880165 | standard |
| 7.20572 | 0.0963776 | standard |
| 7.20981 | 0.0963776 | standard |
| 7.21727 | 0.0880165 | standard |
| 7.84574 | 0.11465 | standard |
| 7.84723 | 0.115313 | standard |
| 7.85733 | 0.115313 | standard |
| 7.85881 | 0.11465 | standard |
| 8.40567 | 0.117542 | standard |
| 8.40718 | 0.120149 | standard |
| 8.413 | 0.118211 | standard |
| 8.41467 | 0.115322 | standard |
| 2.4858 | 1.0 | standard |
| 7.19885 | 0.0874852 | standard |
| 7.20592 | 0.0967999 | standard |
| 7.20961 | 0.0968098 | standard |
| 7.21668 | 0.0879332 | standard |
| 7.84607 | 0.111575 | standard |
| 7.84769 | 0.11236 | standard |
| 7.85686 | 0.111932 | standard |
| 7.85849 | 0.111428 | standard |
| 8.40593 | 0.116463 | standard |
| 8.4074 | 0.119122 | standard |
| 8.4129 | 0.118711 | standard |
| 8.41438 | 0.116309 | standard |
| 2.4858 | 1.0 | standard |
| 7.19944 | 0.0879874 | standard |
| 7.20602 | 0.0966679 | standard |
| 7.20951 | 0.0966098 | standard |
| 7.21618 | 0.0878779 | standard |
| 7.84647 | 0.113102 | standard |
| 7.84788 | 0.113811 | standard |
| 7.85667 | 0.113811 | standard |
| 7.85808 | 0.113102 | standard |
| 8.4062 | 0.115938 | standard |
| 8.40762 | 0.118778 | standard |
| 8.41268 | 0.118778 | standard |
| 8.4141 | 0.115938 | standard |
| 2.4858 | 1.0 | standard |
| 7.20033 | 0.0879452 | standard |
| 7.20621 | 0.096648 | standard |
| 7.20932 | 0.096643 | standard |
| 7.21519 | 0.0877063 | standard |
| 7.84714 | 0.11283 | standard |
| 7.84841 | 0.113559 | standard |
| 7.85615 | 0.113476 | standard |
| 7.85742 | 0.1126 | standard |
| 8.40671 | 0.118568 | standard |
| 8.40781 | 0.121075 | standard |
| 8.41236 | 0.118418 | standard |
| 8.41364 | 0.115425 | standard |
| 2.4858 | 1.0 | standard |
| 7.20073 | 0.0879151 | standard |
| 7.20631 | 0.0962825 | standard |
| 7.20932 | 0.0963547 | standard |
| 7.2148 | 0.0880457 | standard |
| 7.84743 | 0.113132 | standard |
| 7.84861 | 0.113855 | standard |
| 7.85594 | 0.113855 | standard |
| 7.85713 | 0.113132 | standard |
| 8.40685 | 0.116056 | standard |
| 8.40805 | 0.118959 | standard |
| 8.41225 | 0.118959 | standard |
| 8.41345 | 0.116056 | standard |
| 2.4858 | 1.0 | standard |
| 7.20113 | 0.0879543 | standard |
| 7.20641 | 0.0965447 | standard |
| 7.20922 | 0.0965447 | standard |
| 7.2145 | 0.0879543 | standard |
| 7.84762 | 0.113712 | standard |
| 7.84871 | 0.114404 | standard |
| 7.85574 | 0.114404 | standard |
| 7.85683 | 0.113712 | standard |
| 8.40691 | 0.113629 | standard |
| 8.40818 | 0.116819 | standard |
| 8.41212 | 0.116819 | standard |
| 8.41339 | 0.113629 | standard |
| 2.4858 | 1.0 | standard |
| 7.20271 | 0.0880739 | standard |
| 7.20669 | 0.0961643 | standard |
| 7.20893 | 0.0961643 | standard |
| 7.21291 | 0.0880739 | standard |
| 7.84865 | 0.112011 | standard |
| 7.84956 | 0.112776 | standard |
| 7.85489 | 0.112775 | standard |
| 7.85581 | 0.112011 | standard |
| 8.40767 | 0.114634 | standard |
| 8.4086 | 0.117635 | standard |
| 8.4117 | 0.117635 | standard |
| 8.41263 | 0.114634 | standard |