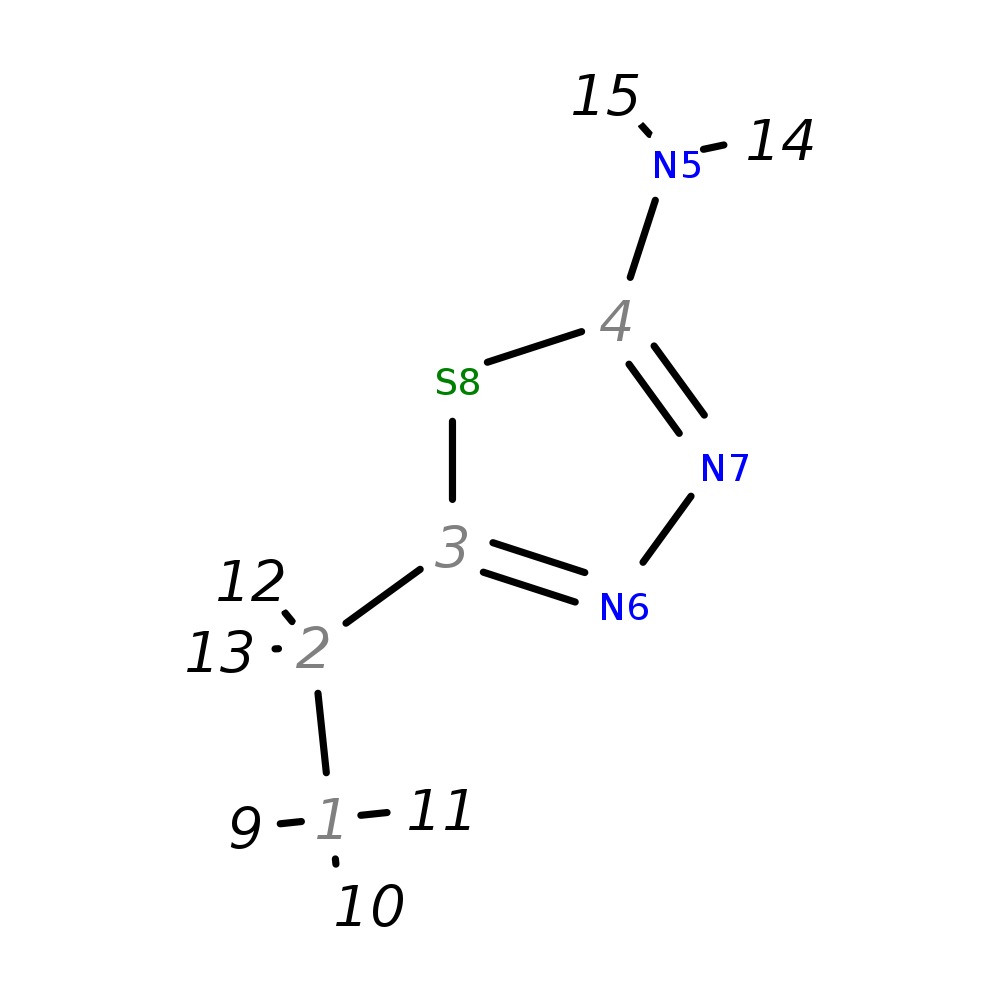
5-Ethyl-1,3,4-thiadiazol-2-amine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01952 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C4H7N3S/c1-2-3-6-7-4(5)8-3/h2H2,1H3,(H2,5,7) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 5-Ethyl-1,3,4-thiadiazol-2-amine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|
| 9 | 1.274 | -14.0 | -14.0 | 8.181 | 8.181 |
| 10 | 0 | 1.274 | -14.0 | 8.181 | 8.181 |
| 11 | 0 | 0 | 1.274 | 8.181 | 8.181 |
| 12 | 0 | 0 | 0 | 2.89 | -14.0 |
| 13 | 0 | 0 | 0 | 0 | 2.89 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.26014 | 0.508612 | standard |
| 1.27376 | 1.00006 | standard |
| 1.28737 | 0.508528 | standard |
| 2.87001 | 0.173975 | standard |
| 2.88364 | 0.501347 | standard |
| 2.89727 | 0.505589 | standard |
| 2.9109 | 0.173587 | standard |
| 1.06208 | 0.372012 | standard |
| 1.2664 | 1.0 | standard |
| 1.46116 | 0.645305 | standard |
| 2.59654 | 0.226362 | standard |
| 2.61692 | 0.223425 | standard |
| 2.79712 | 0.526165 | standard |
| 3.00291 | 0.422044 | standard |
| 3.19702 | 0.108931 | standard |
| 3.22761 | 0.102953 | standard |
| 1.13344 | 0.40532 | standard |
| 1.26877 | 1.0 | standard |
| 1.40319 | 0.580837 | standard |
| 2.69565 | 0.205728 | standard |
| 2.82781 | 0.502973 | standard |
| 2.96402 | 0.436742 | standard |
| 3.10041 | 0.124259 | standard |
| 1.16893 | 0.432161 | standard |
| 1.27072 | 1.00001 | standard |
| 1.37235 | 0.562297 | standard |
| 2.74244 | 0.198839 | standard |
| 2.8432 | 0.505138 | standard |
| 2.9451 | 0.450572 | standard |
| 3.04784 | 0.136643 | standard |
| 1.18075 | 0.440432 | standard |
| 1.27134 | 1.0 | standard |
| 1.3618 | 0.555218 | standard |
| 2.75824 | 0.196042 | standard |
| 2.84826 | 0.505659 | standard |
| 2.9389 | 0.453241 | standard |
| 3.03001 | 0.141049 | standard |
| 1.19024 | 0.444824 | standard |
| 1.2718 | 1.00001 | standard |
| 1.35331 | 0.550496 | standard |
| 2.77108 | 0.193696 | standard |
| 2.85229 | 0.50719 | standard |
| 2.93383 | 0.461033 | standard |
| 3.01579 | 0.144158 | standard |
| 1.23241 | 0.482943 | standard |
| 1.2733 | 1.00009 | standard |
| 1.31415 | 0.528447 | standard |
| 2.82986 | 0.185173 | standard |
| 2.8707 | 0.506626 | standard |
| 2.91155 | 0.48797 | standard |
| 2.95243 | 0.159855 | standard |
| 1.24634 | 0.503733 | standard |
| 1.27359 | 1.00021 | standard |
| 1.30083 | 0.511994 | standard |
| 2.84989 | 0.177108 | standard |
| 2.87709 | 0.493699 | standard |
| 2.90434 | 0.503111 | standard |
| 2.93156 | 0.168943 | standard |
| 1.25322 | 0.504488 | standard |
| 1.27369 | 1.00038 | standard |
| 1.29411 | 0.511681 | standard |
| 2.85995 | 0.173269 | standard |
| 2.88035 | 0.499995 | standard |
| 2.90078 | 0.504084 | standard |
| 2.92121 | 0.173291 | standard |
| 1.25739 | 0.507953 | standard |
| 1.27374 | 1.0 | standard |
| 1.29008 | 0.506066 | standard |
| 2.86594 | 0.17312 | standard |
| 2.8823 | 0.499126 | standard |
| 2.89866 | 0.504019 | standard |
| 2.91502 | 0.173176 | standard |
| 1.26014 | 0.508633 | standard |
| 1.27376 | 1.00006 | standard |
| 1.2874 | 0.508578 | standard |
| 2.87001 | 0.173983 | standard |
| 2.88364 | 0.501334 | standard |
| 2.89727 | 0.505614 | standard |
| 2.9109 | 0.173595 | standard |
| 1.26207 | 0.509755 | standard |
| 1.27379 | 1.00117 | standard |
| 1.28544 | 0.5092 | standard |
| 2.87288 | 0.173804 | standard |
| 2.88458 | 0.50282 | standard |
| 2.89628 | 0.502821 | standard |
| 2.90798 | 0.173805 | standard |
| 1.26286 | 0.509888 | standard |
| 1.27379 | 1.00134 | standard |
| 1.28465 | 0.507921 | standard |
| 2.87407 | 0.173868 | standard |
| 2.88496 | 0.503751 | standard |
| 2.89587 | 0.501135 | standard |
| 2.90679 | 0.173791 | standard |
| 1.26356 | 0.510031 | standard |
| 1.27379 | 1.00152 | standard |
| 1.28398 | 0.510172 | standard |
| 2.87506 | 0.173847 | standard |
| 2.88531 | 0.503095 | standard |
| 2.89552 | 0.501708 | standard |
| 2.9058 | 0.173779 | standard |
| 1.2647 | 0.510326 | standard |
| 1.27379 | 1.00191 | standard |
| 1.28289 | 0.510326 | standard |
| 2.87675 | 0.173931 | standard |
| 2.88585 | 0.504097 | standard |
| 2.89496 | 0.504454 | standard |
| 2.90401 | 0.17402 | standard |
| 1.26519 | 0.510502 | standard |
| 1.27379 | 1.00212 | standard |
| 1.28239 | 0.510502 | standard |
| 2.87749 | 0.174394 | standard |
| 2.88608 | 0.504206 | standard |
| 2.89471 | 0.504197 | standard |
| 2.90332 | 0.173991 | standard |
| 1.26561 | 0.511531 | standard |
| 1.27379 | 1.00234 | standard |
| 1.28195 | 0.510503 | standard |
| 2.87814 | 0.174032 | standard |
| 2.88632 | 0.505461 | standard |
| 2.89448 | 0.503924 | standard |
| 2.90267 | 0.174409 | standard |
| 1.2675 | 0.513217 | standard |
| 1.27379 | 1.00387 | standard |
| 1.28009 | 0.513217 | standard |
| 2.88096 | 0.175055 | standard |
| 2.88724 | 0.505229 | standard |
| 2.89355 | 0.507114 | standard |
| 2.8998 | 0.17514 | standard |