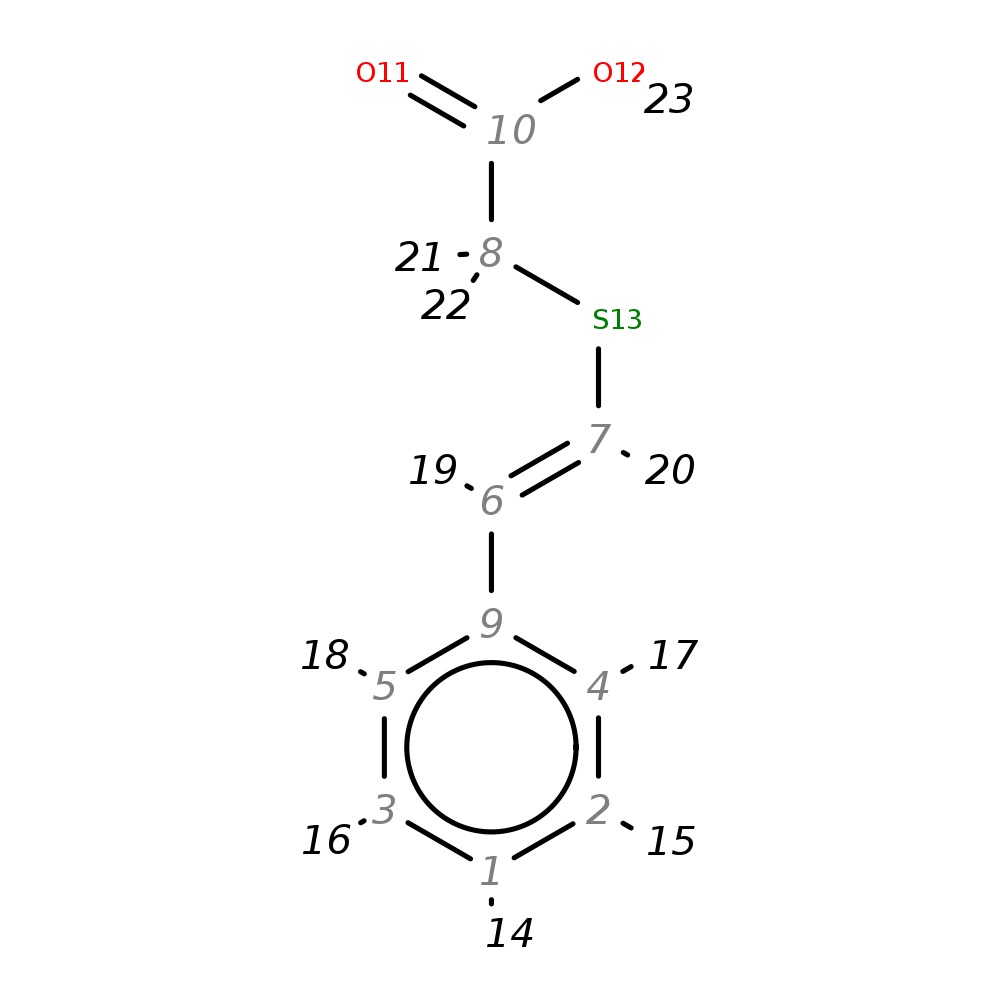
2-(Styrylthio)acetic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02321 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H10O2S/c11-10(12)8-13-7-6-9-4-2-1-3-5-9/h1-7H,8H2,(H,11,12)/b7-6+ | |
| Note 1 | 19?20 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(Styrylthio)acetic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 7.315 | 7.748 | 7.748 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 7.45 | 1.5 | 7.748 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 7.45 | 0 | 7.748 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.534 | 1.5 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 7.534 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 6.397 | 11.16 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 6.574 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.483 | -14.0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.483 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.48302 | 1.0 | standard |
| 6.3872 | 0.226067 | standard |
| 6.40584 | 0.277998 | standard |
| 6.56494 | 0.278019 | standard |
| 6.58348 | 0.226094 | standard |
| 7.3019 | 0.107329 | standard |
| 7.31472 | 0.23355 | standard |
| 7.32748 | 0.154402 | standard |
| 7.43728 | 0.207523 | standard |
| 7.45003 | 0.416641 | standard |
| 7.46286 | 0.22672 | standard |
| 7.52835 | 0.485885 | standard |
| 7.5411 | 0.363764 | standard |
| 3.48302 | 0.868652 | standard |
| 6.18104 | 0.0400521 | standard |
| 6.46181 | 0.481863 | standard |
| 6.50888 | 0.482005 | standard |
| 6.78974 | 0.0423561 | standard |
| 7.45619 | 1.0 | standard |
| 3.48302 | 1.0 | standard |
| 6.26421 | 0.074585 | standard |
| 6.45028 | 0.455615 | standard |
| 6.5204 | 0.455724 | standard |
| 6.70658 | 0.075355 | standard |
| 7.14523 | 0.0176431 | standard |
| 7.3437 | 0.248569 | standard |
| 7.39417 | 0.312382 | standard |
| 7.45226 | 0.710822 | standard |
| 7.49414 | 0.715186 | standard |
| 3.48302 | 1.0 | standard |
| 6.30304 | 0.0999666 | standard |
| 6.44258 | 0.414775 | standard |
| 6.5281 | 0.414851 | standard |
| 6.66764 | 0.100312 | standard |
| 7.19492 | 0.0223401 | standard |
| 7.22398 | 0.0347541 | standard |
| 7.29277 | 0.095106 | standard |
| 7.30244 | 0.0947561 | standard |
| 7.37987 | 0.280726 | standard |
| 7.4489 | 0.513539 | standard |
| 7.50527 | 0.665104 | standard |
| 7.58423 | 0.121957 | standard |
| 3.48302 | 1.0 | standard |
| 6.31542 | 0.110926 | standard |
| 6.43939 | 0.40068 | standard |
| 6.5313 | 0.400744 | standard |
| 6.65526 | 0.111179 | standard |
| 7.21327 | 0.0259281 | standard |
| 7.23611 | 0.037598 | standard |
| 7.29448 | 0.0990761 | standard |
| 7.37487 | 0.301585 | standard |
| 7.44842 | 0.462524 | standard |
| 7.4772 | 0.293409 | standard |
| 7.5074 | 0.675075 | standard |
| 7.58069 | 0.123721 | standard |
| 3.48302 | 1.0 | standard |
| 6.32503 | 0.120743 | standard |
| 6.4367 | 0.388914 | standard |
| 6.53408 | 0.38897 | standard |
| 6.64565 | 0.120941 | standard |
| 7.24109 | 0.036409 | standard |
| 7.29706 | 0.10532 | standard |
| 7.3271 | 0.111408 | standard |
| 7.35533 | 0.192218 | standard |
| 7.37603 | 0.274082 | standard |
| 7.44838 | 0.424379 | standard |
| 7.50883 | 0.690819 | standard |
| 7.57727 | 0.129082 | standard |
| 3.48302 | 1.0 | standard |
| 6.36479 | 0.177675 | standard |
| 6.42062 | 0.327332 | standard |
| 6.55016 | 0.327355 | standard |
| 6.60589 | 0.17772 | standard |
| 7.27367 | 0.0700813 | standard |
| 7.31033 | 0.163463 | standard |
| 7.34584 | 0.180728 | standard |
| 7.41188 | 0.195025 | standard |
| 7.44919 | 0.37322 | standard |
| 7.48583 | 0.248273 | standard |
| 7.51961 | 0.574464 | standard |
| 7.55704 | 0.233248 | standard |
| 3.48302 | 1.0 | standard |
| 6.37649 | 0.201078 | standard |
| 6.41368 | 0.303334 | standard |
| 6.5571 | 0.303343 | standard |
| 6.59429 | 0.201092 | standard |
| 7.28787 | 0.0872775 | standard |
| 7.31308 | 0.195504 | standard |
| 7.3378 | 0.175299 | standard |
| 7.42436 | 0.197732 | standard |
| 7.44955 | 0.391413 | standard |
| 7.47476 | 0.235243 | standard |
| 7.52344 | 0.53502 | standard |
| 7.54884 | 0.296278 | standard |
| 3.48302 | 1.0 | standard |
| 6.38194 | 0.213421 | standard |
| 6.40981 | 0.290805 | standard |
| 6.56087 | 0.29081 | standard |
| 6.58874 | 0.213428 | standard |
| 7.29494 | 0.0967724 | standard |
| 7.31401 | 0.213773 | standard |
| 7.33293 | 0.165579 | standard |
| 7.43081 | 0.202948 | standard |
| 7.4498 | 0.407666 | standard |
| 7.46892 | 0.231687 | standard |
| 7.52576 | 0.511939 | standard |
| 7.54487 | 0.329307 | standard |
| 3.48302 | 1.0 | standard |
| 6.38511 | 0.220981 | standard |
| 6.40743 | 0.283151 | standard |
| 6.56325 | 0.283155 | standard |
| 6.58557 | 0.220986 | standard |
| 7.29913 | 0.103007 | standard |
| 7.31447 | 0.225806 | standard |
| 7.32976 | 0.159068 | standard |
| 7.43469 | 0.205259 | standard |
| 7.44995 | 0.414985 | standard |
| 7.46531 | 0.22926 | standard |
| 7.52729 | 0.496905 | standard |
| 7.54259 | 0.350464 | standard |
| 3.48302 | 1.0 | standard |
| 6.3872 | 0.226067 | standard |
| 6.40584 | 0.277998 | standard |
| 6.56494 | 0.27802 | standard |
| 6.58348 | 0.226093 | standard |
| 7.3019 | 0.107329 | standard |
| 7.31472 | 0.233551 | standard |
| 7.32748 | 0.154402 | standard |
| 7.43728 | 0.207524 | standard |
| 7.45003 | 0.416642 | standard |
| 7.46286 | 0.226721 | standard |
| 7.52835 | 0.485887 | standard |
| 7.5411 | 0.363765 | standard |
| 3.48302 | 1.0 | standard |
| 6.38868 | 0.229735 | standard |
| 6.40465 | 0.27431 | standard |
| 6.56613 | 0.274311 | standard |
| 6.5821 | 0.229737 | standard |
| 7.30388 | 0.110193 | standard |
| 7.31488 | 0.237569 | standard |
| 7.32589 | 0.150595 | standard |
| 7.43915 | 0.210092 | standard |
| 7.45006 | 0.419006 | standard |
| 7.46108 | 0.226434 | standard |
| 7.52913 | 0.478178 | standard |
| 7.54005 | 0.371665 | standard |
| 3.48302 | 1.0 | standard |
| 6.38928 | 0.231213 | standard |
| 6.40416 | 0.272836 | standard |
| 6.56662 | 0.272837 | standard |
| 6.5815 | 0.231215 | standard |
| 7.30465 | 0.111513 | standard |
| 7.31494 | 0.240401 | standard |
| 7.32522 | 0.149331 | standard |
| 7.43989 | 0.211599 | standard |
| 7.45009 | 0.422979 | standard |
| 7.46034 | 0.22691 | standard |
| 7.52943 | 0.475188 | standard |
| 7.53965 | 0.377116 | standard |
| 3.48302 | 1.0 | standard |
| 6.38978 | 0.232521 | standard |
| 6.40366 | 0.271555 | standard |
| 6.56702 | 0.271531 | standard |
| 6.581 | 0.232492 | standard |
| 7.30535 | 0.112559 | standard |
| 7.31499 | 0.242357 | standard |
| 7.32463 | 0.148038 | standard |
| 7.44052 | 0.209794 | standard |
| 7.4501 | 0.422368 | standard |
| 7.45975 | 0.225838 | standard |
| 7.52972 | 0.472333 | standard |
| 7.53929 | 0.379064 | standard |
| 3.48302 | 1.0 | standard |
| 6.39057 | 0.234654 | standard |
| 6.40297 | 0.269375 | standard |
| 6.56772 | 0.269376 | standard |
| 6.58011 | 0.234655 | standard |
| 7.30648 | 0.114292 | standard |
| 7.31507 | 0.244576 | standard |
| 7.32363 | 0.145886 | standard |
| 7.4416 | 0.213005 | standard |
| 7.45015 | 0.423973 | standard |
| 7.45869 | 0.224844 | standard |
| 7.53018 | 0.467 | standard |
| 7.53871 | 0.384467 | standard |
| 3.48302 | 1.0 | standard |
| 6.39097 | 0.235576 | standard |
| 6.40267 | 0.268476 | standard |
| 6.56801 | 0.268446 | standard |
| 6.57981 | 0.235542 | standard |
| 7.30696 | 0.115224 | standard |
| 7.31509 | 0.245144 | standard |
| 7.32321 | 0.145192 | standard |
| 7.4421 | 0.213541 | standard |
| 7.45016 | 0.422724 | standard |
| 7.45827 | 0.22512 | standard |
| 7.53038 | 0.464316 | standard |
| 7.53845 | 0.387679 | standard |
| 3.48302 | 1.0 | standard |
| 6.39126 | 0.236397 | standard |
| 6.40237 | 0.267658 | standard |
| 6.56831 | 0.267626 | standard |
| 6.57952 | 0.236361 | standard |
| 7.30738 | 0.115712 | standard |
| 7.31509 | 0.245979 | standard |
| 7.32282 | 0.144402 | standard |
| 7.44249 | 0.211861 | standard |
| 7.45017 | 0.422762 | standard |
| 7.45788 | 0.225051 | standard |
| 7.53055 | 0.464474 | standard |
| 7.53821 | 0.390684 | standard |
| 3.48302 | 1.0 | standard |
| 6.39255 | 0.239947 | standard |
| 6.40118 | 0.264017 | standard |
| 6.5695 | 0.264017 | standard |
| 6.57813 | 0.239948 | standard |
| 7.30924 | 0.118908 | standard |
| 7.31521 | 0.249429 | standard |
| 7.32113 | 0.140917 | standard |
| 7.44426 | 0.213118 | standard |
| 7.45019 | 0.424461 | standard |
| 7.45613 | 0.222857 | standard |
| 7.53139 | 0.45566 | standard |
| 7.53731 | 0.398801 | standard |