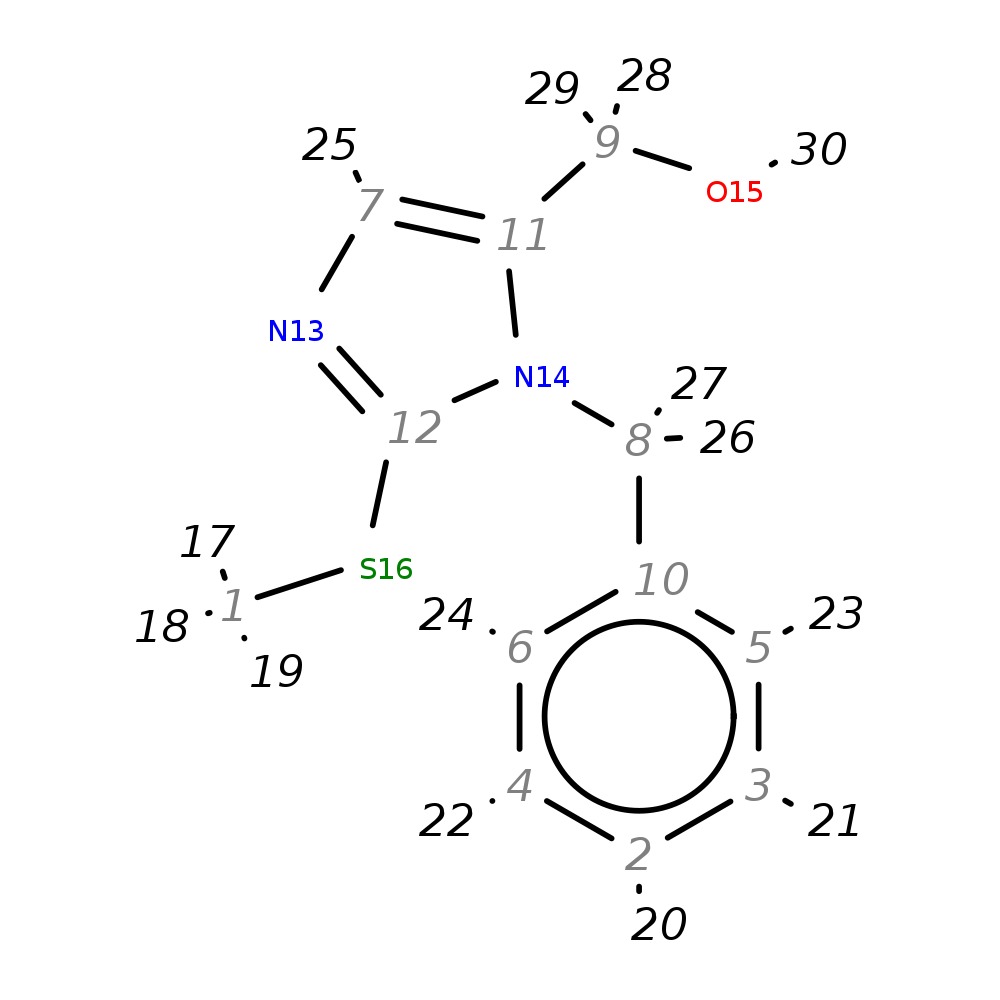
[1-Benzyl-2-(methylsulfanyl)-1H-imidazol-5-yl]methanol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01653 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H14N2OS/c1-16-12-13-7-11(9-15)14(12)8-10-5-3-2-4-6-10/h2-7,15H,8-9H2,1H3 | |
| Note 1 | 26,27 weak intensity (saturated) | |
| Note 2 | 28,29?26,27 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| [1-Benzyl-2-(methylsulfanyl)-1H-imidazol-5-yl]methanol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 2.443 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 2.443 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 2.443 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 7.347 | 7.765 | 7.765 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 7.392 | 1.5 | 8.227 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 7.392 | 0 | 8.227 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 7.092 | 1.5 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.092 | 0 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.138 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.345 | -14.0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.345 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.521 | -14.0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.521 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666654 | standard |
| 5.34485 | 0.666933 | standard |
| 7.08498 | 0.260323 | standard |
| 7.09819 | 0.283229 | standard |
| 7.13814 | 0.33396 | standard |
| 7.33247 | 0.0495047 | standard |
| 7.34523 | 0.106902 | standard |
| 7.3578 | 0.142679 | standard |
| 7.37993 | 0.185852 | standard |
| 7.39308 | 0.229016 | standard |
| 7.40573 | 0.0875123 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.667318 | standard |
| 5.34485 | 0.667417 | standard |
| 6.98147 | 0.0909841 | standard |
| 7.14129 | 0.530612 | standard |
| 7.22805 | 0.274689 | standard |
| 7.26088 | 0.280908 | standard |
| 7.32742 | 0.50335 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.667223 | standard |
| 5.34485 | 0.667037 | standard |
| 7.02739 | 0.122112 | standard |
| 7.13974 | 0.537034 | standard |
| 7.34113 | 0.438477 | standard |
| 7.41422 | 0.165897 | standard |
| 7.57937 | 0.0135681 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.66715 | standard |
| 5.34485 | 0.667128 | standard |
| 7.04512 | 0.138048 | standard |
| 7.13731 | 0.533045 | standard |
| 7.22904 | 0.0423651 | standard |
| 7.34927 | 0.433542 | standard |
| 7.39835 | 0.172022 | standard |
| 7.42073 | 0.170808 | standard |
| 7.5392 | 0.0161081 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666962 | standard |
| 5.34485 | 0.667085 | standard |
| 7.05031 | 0.143819 | standard |
| 7.13687 | 0.503657 | standard |
| 7.23995 | 0.0305901 | standard |
| 7.35208 | 0.425152 | standard |
| 7.41653 | 0.174359 | standard |
| 7.466 | 0.0491041 | standard |
| 7.52164 | 0.0175393 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666395 | standard |
| 5.34485 | 0.666775 | standard |
| 7.05415 | 0.149214 | standard |
| 7.13738 | 0.475181 | standard |
| 7.25324 | 0.0224511 | standard |
| 7.32039 | 0.103866 | standard |
| 7.35463 | 0.413119 | standard |
| 7.41298 | 0.176948 | standard |
| 7.45885 | 0.0498303 | standard |
| 7.50755 | 0.0190063 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666623 | standard |
| 5.34485 | 0.667054 | standard |
| 7.07204 | 0.190887 | standard |
| 7.11072 | 0.221482 | standard |
| 7.1381 | 0.352575 | standard |
| 7.29936 | 0.0202346 | standard |
| 7.34914 | 0.109535 | standard |
| 7.36613 | 0.403243 | standard |
| 7.39888 | 0.181234 | standard |
| 7.42784 | 0.0504733 | standard |
| 7.44473 | 0.033596 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666386 | standard |
| 5.34485 | 0.666485 | standard |
| 7.07865 | 0.224704 | standard |
| 7.10398 | 0.250682 | standard |
| 7.13813 | 0.339934 | standard |
| 7.316 | 0.0290299 | standard |
| 7.34176 | 0.068566 | standard |
| 7.34659 | 0.0717027 | standard |
| 7.36432 | 0.227053 | standard |
| 7.37124 | 0.259415 | standard |
| 7.39553 | 0.187541 | standard |
| 7.41706 | 0.0576492 | standard |
| 7.42413 | 0.0468816 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666543 | standard |
| 5.34485 | 0.666757 | standard |
| 7.08169 | 0.246249 | standard |
| 7.10104 | 0.272831 | standard |
| 7.13813 | 0.335606 | standard |
| 7.32433 | 0.0376955 | standard |
| 7.34339 | 0.0825191 | standard |
| 7.3614 | 0.170184 | standard |
| 7.37531 | 0.210003 | standard |
| 7.3942 | 0.200883 | standard |
| 7.41151 | 0.0664257 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666715 | standard |
| 5.34485 | 0.666919 | standard |
| 7.0836 | 0.2543 | standard |
| 7.09931 | 0.281962 | standard |
| 7.13813 | 0.334245 | standard |
| 7.32926 | 0.0443511 | standard |
| 7.34458 | 0.095848 | standard |
| 7.3594 | 0.154121 | standard |
| 7.37796 | 0.19571 | standard |
| 7.39351 | 0.215842 | standard |
| 7.40825 | 0.0767893 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.6667 | standard |
| 5.34485 | 0.666587 | standard |
| 7.08498 | 0.26029 | standard |
| 7.09819 | 0.283195 | standard |
| 7.13814 | 0.333917 | standard |
| 7.33247 | 0.0494976 | standard |
| 7.34523 | 0.106889 | standard |
| 7.3578 | 0.142661 | standard |
| 7.37993 | 0.18583 | standard |
| 7.39308 | 0.228987 | standard |
| 7.40573 | 0.0875013 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.667314 | standard |
| 5.34485 | 0.667254 | standard |
| 7.0859 | 0.26439 | standard |
| 7.09732 | 0.283044 | standard |
| 7.13814 | 0.33399 | standard |
| 7.33472 | 0.0535022 | standard |
| 7.34575 | 0.115859 | standard |
| 7.35653 | 0.134329 | standard |
| 7.38143 | 0.178854 | standard |
| 7.39275 | 0.238004 | standard |
| 7.4037 | 0.0942681 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.665553 | standard |
| 5.34485 | 0.666057 | standard |
| 7.08629 | 0.264759 | standard |
| 7.09699 | 0.284429 | standard |
| 7.13814 | 0.333487 | standard |
| 7.33561 | 0.0550691 | standard |
| 7.34587 | 0.118967 | standard |
| 7.35602 | 0.130831 | standard |
| 7.38206 | 0.177007 | standard |
| 7.39263 | 0.244814 | standard |
| 7.4029 | 0.0994786 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666681 | standard |
| 5.34485 | 0.666571 | standard |
| 7.08668 | 0.265923 | standard |
| 7.09665 | 0.284869 | standard |
| 7.13814 | 0.33366 | standard |
| 7.33632 | 0.0567597 | standard |
| 7.346 | 0.122957 | standard |
| 7.35552 | 0.127636 | standard |
| 7.38262 | 0.175093 | standard |
| 7.39252 | 0.247463 | standard |
| 7.40216 | 0.101546 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666302 | standard |
| 5.34485 | 0.6671 | standard |
| 7.08721 | 0.266658 | standard |
| 7.09616 | 0.286864 | standard |
| 7.13814 | 0.333594 | standard |
| 7.33761 | 0.0593823 | standard |
| 7.34617 | 0.12968 | standard |
| 7.35465 | 0.122878 | standard |
| 7.38353 | 0.170153 | standard |
| 7.39236 | 0.251931 | standard |
| 7.401 | 0.105383 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.66639 | standard |
| 5.34485 | 0.666531 | standard |
| 7.08745 | 0.270194 | standard |
| 7.09593 | 0.285857 | standard |
| 7.13814 | 0.333181 | standard |
| 7.33813 | 0.0605735 | standard |
| 7.34627 | 0.131161 | standard |
| 7.35436 | 0.120493 | standard |
| 7.38397 | 0.171134 | standard |
| 7.39234 | 0.25372 | standard |
| 7.40052 | 0.110845 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.666592 | standard |
| 5.34485 | 0.666235 | standard |
| 7.08765 | 0.273971 | standard |
| 7.09573 | 0.280655 | standard |
| 7.13814 | 0.333588 | standard |
| 7.33862 | 0.0616603 | standard |
| 7.34634 | 0.13417 | standard |
| 7.35404 | 0.11887 | standard |
| 7.38433 | 0.168822 | standard |
| 7.39223 | 0.256831 | standard |
| 7.40009 | 0.113449 | standard |
| 2.44315 | 1.0 | standard |
| 4.5211 | 0.667085 | standard |
| 5.34485 | 0.667381 | standard |
| 7.08857 | 0.27737 | standard |
| 7.09485 | 0.282582 | standard |
| 7.13814 | 0.33368 | standard |
| 7.34065 | 0.06657 | standard |
| 7.34661 | 0.144984 | standard |
| 7.35255 | 0.110616 | standard |
| 7.38595 | 0.16153 | standard |
| 7.39207 | 0.264186 | standard |
| 7.39811 | 0.119533 | standard |