4'-Methoxyacetophenone
Simulation outputs:
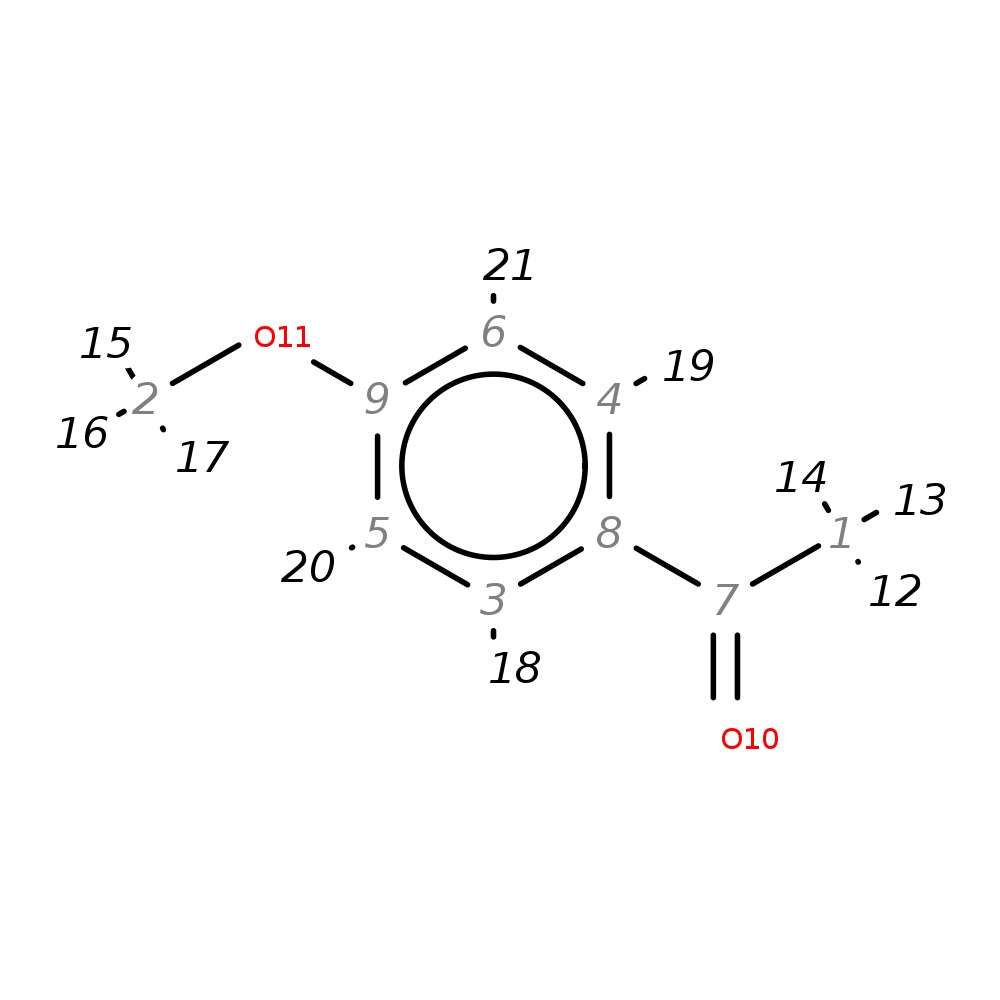
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01893 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H10O2/c1-7(10)8-3-5-9(11-2)6-4-8/h3-6H,1-2H3 | |
| Note 1 | 20,21?18,19 | |
| Note 2 | 12,13,14?15,16,17 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4'-Methoxyacetophenone | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 2.618 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 2.618 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 2.618 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 3.911 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 3.911 | -14.0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 3.911 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 8.018 | 1.5 | 9.39 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.018 | 0 | 9.39 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.092 | 1.5 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.092 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.999538 | standard |
| 7.08394 | 0.282232 | standard |
| 7.0995 | 0.282781 | standard |
| 8.01033 | 0.282792 | standard |
| 8.0259 | 0.280998 | standard |
| 2.61822 | 0.998719 | standard |
| 3.91111 | 1.0 | standard |
| 6.9602 | 0.215255 | standard |
| 7.19426 | 0.349291 | standard |
| 7.9155 | 0.349253 | standard |
| 8.14959 | 0.215245 | standard |
| 2.61821 | 0.998833 | standard |
| 3.91111 | 1.0 | standard |
| 7.00744 | 0.236954 | standard |
| 7.16311 | 0.327361 | standard |
| 7.94665 | 0.327305 | standard |
| 8.1024 | 0.236828 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.997471 | standard |
| 7.02978 | 0.247779 | standard |
| 7.14644 | 0.315985 | standard |
| 7.96335 | 0.315925 | standard |
| 8.08005 | 0.247768 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.996958 | standard |
| 7.03701 | 0.251477 | standard |
| 7.14072 | 0.312144 | standard |
| 7.96911 | 0.312141 | standard |
| 8.07283 | 0.251347 | standard |
| 2.61821 | 0.999961 | standard |
| 3.91111 | 1.0 | standard |
| 7.04276 | 0.254854 | standard |
| 7.13609 | 0.309543 | standard |
| 7.97374 | 0.309544 | standard |
| 8.06701 | 0.254803 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.999791 | standard |
| 7.06787 | 0.268117 | standard |
| 7.11447 | 0.296023 | standard |
| 7.99526 | 0.296134 | standard |
| 8.04196 | 0.268099 | standard |
| 2.61821 | 0.99871 | standard |
| 3.91111 | 1.0 | standard |
| 7.07595 | 0.272429 | standard |
| 7.10709 | 0.291109 | standard |
| 8.00275 | 0.291108 | standard |
| 8.03388 | 0.272429 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.999663 | standard |
| 7.08 | 0.279524 | standard |
| 7.10324 | 0.285362 | standard |
| 8.00649 | 0.284734 | standard |
| 8.02984 | 0.2795 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.998509 | standard |
| 7.08236 | 0.279501 | standard |
| 7.10097 | 0.284543 | standard |
| 8.00875 | 0.283456 | standard |
| 8.02748 | 0.279468 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.998811 | standard |
| 7.08394 | 0.282131 | standard |
| 7.0995 | 0.282679 | standard |
| 8.01033 | 0.282691 | standard |
| 8.0259 | 0.280897 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.99929 | standard |
| 7.08512 | 0.282211 | standard |
| 7.09841 | 0.282676 | standard |
| 8.01142 | 0.282676 | standard |
| 8.02471 | 0.28221 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.999683 | standard |
| 7.08551 | 0.282341 | standard |
| 7.09792 | 0.281901 | standard |
| 8.01181 | 0.282281 | standard |
| 8.02432 | 0.282281 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.998471 | standard |
| 7.0859 | 0.281865 | standard |
| 7.09753 | 0.281865 | standard |
| 8.01221 | 0.281802 | standard |
| 8.02393 | 0.282293 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.999287 | standard |
| 7.0866 | 0.28216 | standard |
| 7.09694 | 0.28269 | standard |
| 8.0129 | 0.28216 | standard |
| 8.02324 | 0.28269 | standard |
| 2.61821 | 0.999912 | standard |
| 3.91111 | 1.0 | standard |
| 7.08689 | 0.282791 | standard |
| 7.09664 | 0.282791 | standard |
| 8.01319 | 0.282791 | standard |
| 8.02294 | 0.282791 | standard |
| 2.61821 | 1.0 | standard |
| 3.91111 | 0.998354 | standard |
| 7.08708 | 0.281761 | standard |
| 7.09645 | 0.281761 | standard |
| 8.01339 | 0.281163 | standard |
| 8.02275 | 0.281757 | standard |
| 2.61821 | 0.998779 | standard |
| 3.91111 | 1.0 | standard |
| 7.08817 | 0.281895 | standard |
| 7.09536 | 0.281895 | standard |
| 8.01447 | 0.281122 | standard |
| 8.02166 | 0.28189 | standard |