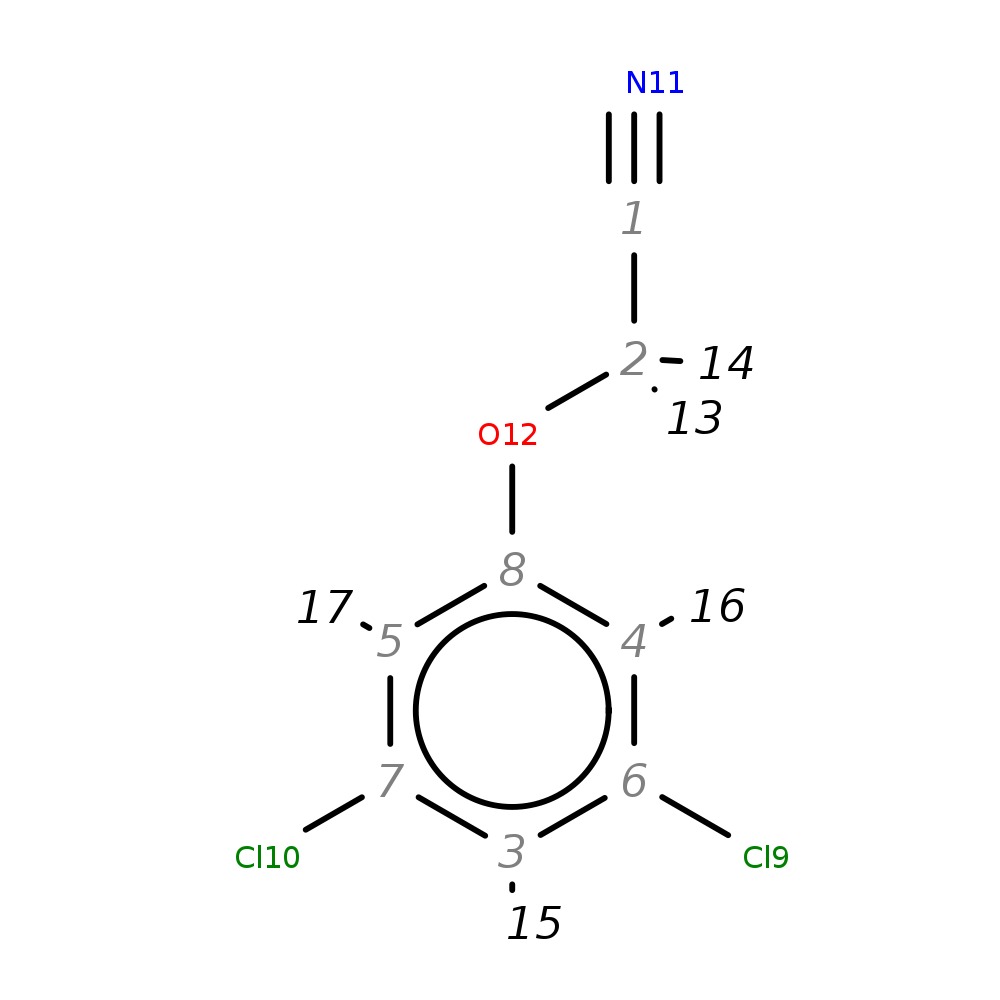
2-(3,5-Dichlorophenoxy)acetonitrile
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02188 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H5Cl2NO/c9-6-3-7(10)5-8(4-6)12-2-1-11/h3-5H,2H2 | |
| Note 1 | Cl causing isotope shifts for aromatics lineshapes) | |
| Note 2 | 13,14 weak inmtensity (saturated) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(3,5-Dichlorophenoxy)acetonitrile | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|
| 13 | 7.124 | -14.0 | 1.906 | 0 | 0 |
| 14 | 0 | 7.124 | 1.906 | 0 | 0 |
| 15 | 0 | 0 | 7.248 | 0.79 | 0.79 |
| 16 | 0 | 0 | 0 | 5.034 | 1.5 |
| 17 | 0 | 0 | 0 | 0 | 5.034 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 5.03385 | 1.00037 | standard |
| 7.12264 | 0.696123 | standard |
| 7.12547 | 0.712814 | standard |
| 7.24787 | 0.305838 | standard |
| 5.03379 | 1.0 | standard |
| 7.14193 | 0.902316 | standard |
| 7.20771 | 0.405192 | standard |
| 5.0338 | 1.0 | standard |
| 7.11157 | 0.606256 | standard |
| 7.13673 | 0.827264 | standard |
| 7.24025 | 0.31613 | standard |
| 5.0338 | 1.0 | standard |
| 7.11361 | 0.627585 | standard |
| 7.13389 | 0.794929 | standard |
| 7.24673 | 0.308785 | standard |
| 5.0338 | 1.0 | standard |
| 7.11463 | 0.635309 | standard |
| 7.13288 | 0.784086 | standard |
| 7.2473 | 0.307085 | standard |
| 5.0338 | 1.0 | standard |
| 7.1155 | 0.642594 | standard |
| 7.13209 | 0.777225 | standard |
| 7.24757 | 0.307044 | standard |
| 5.0338 | 1.0 | standard |
| 7.11971 | 0.674888 | standard |
| 7.12823 | 0.742335 | standard |
| 7.24784 | 0.306232 | standard |
| 5.03381 | 1.0 | standard |
| 7.12116 | 0.682413 | standard |
| 7.12686 | 0.7274 | standard |
| 7.24783 | 0.304387 | standard |
| 5.0338 | 1.0 | standard |
| 7.1219 | 0.692265 | standard |
| 7.12615 | 0.725832 | standard |
| 7.24786 | 0.305825 | standard |
| 5.0338 | 1.0 | standard |
| 7.12234 | 0.694772 | standard |
| 7.12575 | 0.722063 | standard |
| 7.24784 | 0.30556 | standard |
| 5.03385 | 1.00037 | standard |
| 7.12264 | 0.696123 | standard |
| 7.12547 | 0.712815 | standard |
| 7.24787 | 0.30584 | standard |
| 5.03385 | 1.00052 | standard |
| 7.12281 | 0.690614 | standard |
| 7.12531 | 0.705062 | standard |
| 7.24789 | 0.303678 | standard |
| 5.03385 | 1.00051 | standard |
| 7.1229 | 0.705229 | standard |
| 7.12521 | 0.718924 | standard |
| 7.2478 | 0.308372 | standard |
| 5.0338 | 1.0 | standard |
| 7.12299 | 0.706862 | standard |
| 7.12511 | 0.711064 | standard |
| 7.24784 | 0.305928 | standard |
| 5.03385 | 1.00077 | standard |
| 7.12307 | 0.703361 | standard |
| 7.12504 | 0.70718 | standard |
| 7.24782 | 0.307113 | standard |
| 5.0338 | 1.0 | standard |
| 7.12317 | 0.700123 | standard |
| 7.12493 | 0.703588 | standard |
| 7.24782 | 0.303249 | standard |
| 5.0338 | 1.0 | standard |
| 7.12315 | 0.698489 | standard |
| 7.12495 | 0.701932 | standard |
| 7.24784 | 0.307174 | standard |
| 5.0338 | 1.0 | standard |
| 7.12345 | 0.71631 | standard |
| 7.12465 | 0.716311 | standard |
| 7.24784 | 0.303201 | standard |