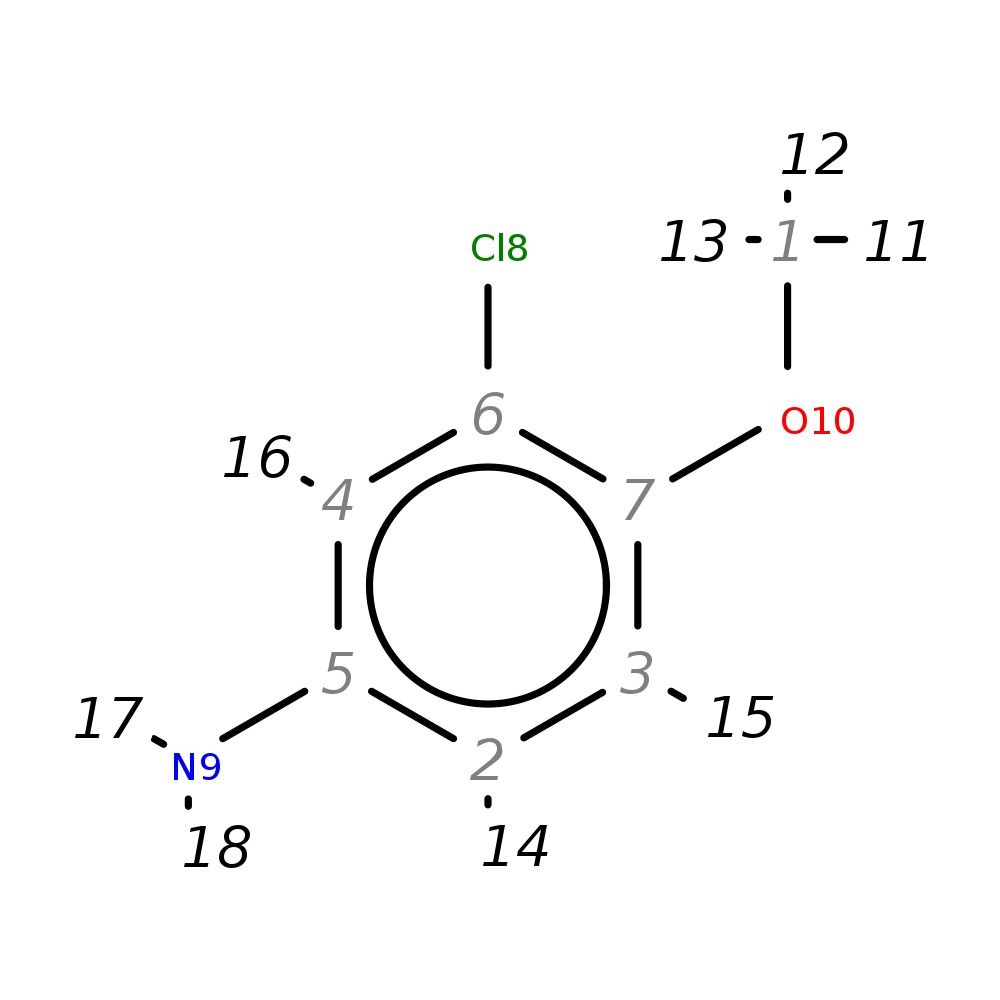
3-Chloro-4-methoxyaniline
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02878 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H8ClNO/c1-10-7-3-2-5(9)4-6(7)8/h2-4H,9H2,1H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Chloro-4-methoxyaniline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|
| 11 | 3.835 | -14.0 | -14.0 | 0 | 0 | 0 |
| 12 | 0 | 3.835 | -14.0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 3.835 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 6.775 | 8.875 | 3.147 |
| 15 | 0 | 0 | 0 | 0 | 7.002 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 6.943 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.83454 | 1.0 | standard |
| 6.76475 | 0.0854683 | standard |
| 6.76992 | 0.090231 | standard |
| 6.77951 | 0.09853 | standard |
| 6.78469 | 0.10027 | standard |
| 6.9402 | 0.185516 | standard |
| 6.94537 | 0.179281 | standard |
| 6.99501 | 0.179891 | standard |
| 7.00979 | 0.158362 | standard |
| 3.83454 | 1.0 | standard |
| 6.58334 | 0.0300461 | standard |
| 6.65168 | 0.0370371 | standard |
| 6.86584 | 0.297003 | standard |
| 6.92743 | 0.487885 | standard |
| 7.15753 | 0.0522941 | standard |
| 3.83454 | 1.0 | standard |
| 6.65336 | 0.0395962 | standard |
| 6.70147 | 0.0521894 | standard |
| 6.7986 | 0.103489 | standard |
| 6.84552 | 0.205739 | standard |
| 6.92648 | 0.314806 | standard |
| 6.95193 | 0.359064 | standard |
| 7.09744 | 0.0790821 | standard |
| 3.83454 | 1.0 | standard |
| 6.68707 | 0.0481872 | standard |
| 6.72393 | 0.063101 | standard |
| 6.7966 | 0.105952 | standard |
| 6.83319 | 0.173311 | standard |
| 6.92723 | 0.253335 | standard |
| 6.96093 | 0.382032 | standard |
| 7.07003 | 0.0982679 | standard |
| 3.83454 | 1.0 | standard |
| 6.69787 | 0.0515972 | standard |
| 6.73097 | 0.066856 | standard |
| 6.79542 | 0.106251 | standard |
| 6.82848 | 0.161763 | standard |
| 6.92838 | 0.239246 | standard |
| 6.96247 | 0.375652 | standard |
| 7.06135 | 0.105174 | standard |
| 3.83454 | 1.0 | standard |
| 6.70643 | 0.054578 | standard |
| 6.73633 | 0.069843 | standard |
| 6.79441 | 0.106121 | standard |
| 6.82423 | 0.15069 | standard |
| 6.92937 | 0.229429 | standard |
| 6.96413 | 0.337791 | standard |
| 7.0546 | 0.110855 | standard |
| 3.83454 | 1.0 | standard |
| 6.74276 | 0.0709366 | standard |
| 6.75808 | 0.0830497 | standard |
| 6.78696 | 0.103027 | standard |
| 6.80227 | 0.119857 | standard |
| 6.93543 | 0.198925 | standard |
| 6.95073 | 0.173683 | standard |
| 6.9821 | 0.205579 | standard |
| 7.02639 | 0.138035 | standard |
| 3.83454 | 1.0 | standard |
| 6.754 | 0.0775472 | standard |
| 6.76425 | 0.0870472 | standard |
| 6.78353 | 0.100465 | standard |
| 6.79377 | 0.110256 | standard |
| 6.93776 | 0.192885 | standard |
| 6.94804 | 0.174819 | standard |
| 6.98832 | 0.192019 | standard |
| 7.01786 | 0.148515 | standard |
| 3.83454 | 1.0 | standard |
| 6.75942 | 0.0813336 | standard |
| 6.76713 | 0.0887626 | standard |
| 6.78162 | 0.0992177 | standard |
| 6.78933 | 0.105645 | standard |
| 6.93893 | 0.189646 | standard |
| 6.94668 | 0.176196 | standard |
| 6.99159 | 0.185783 | standard |
| 7.01375 | 0.153388 | standard |
| 3.83454 | 1.0 | standard |
| 6.76268 | 0.0835587 | standard |
| 6.76883 | 0.090041 | standard |
| 6.78042 | 0.0986543 | standard |
| 6.78656 | 0.102748 | standard |
| 6.93972 | 0.187983 | standard |
| 6.94585 | 0.17772 | standard |
| 6.99362 | 0.182238 | standard |
| 7.01137 | 0.156344 | standard |
| 3.83454 | 1.0 | standard |
| 6.76475 | 0.0854532 | standard |
| 6.76992 | 0.0902211 | standard |
| 6.77951 | 0.0984691 | standard |
| 6.78469 | 0.0995519 | standard |
| 6.9402 | 0.185462 | standard |
| 6.94537 | 0.178632 | standard |
| 6.99501 | 0.179889 | standard |
| 7.00979 | 0.157737 | standard |
| 3.83454 | 1.0 | standard |
| 6.76623 | 0.0867667 | standard |
| 6.77072 | 0.0902598 | standard |
| 6.77891 | 0.0986563 | standard |
| 6.7833 | 0.0987596 | standard |
| 6.94059 | 0.184212 | standard |
| 6.94498 | 0.180877 | standard |
| 6.996 | 0.17825 | standard |
| 7.0086 | 0.159817 | standard |
| 3.83454 | 1.0 | standard |
| 6.76682 | 0.0873936 | standard |
| 6.77102 | 0.0901727 | standard |
| 6.77871 | 0.0982957 | standard |
| 6.78281 | 0.0977529 | standard |
| 6.94069 | 0.183711 | standard |
| 6.94478 | 0.180695 | standard |
| 6.9964 | 0.177563 | standard |
| 7.0082 | 0.159856 | standard |
| 3.83454 | 1.0 | standard |
| 6.76742 | 0.088521 | standard |
| 6.77122 | 0.0909176 | standard |
| 6.77841 | 0.0980211 | standard |
| 6.78232 | 0.0974205 | standard |
| 6.94079 | 0.182895 | standard |
| 6.94469 | 0.181139 | standard |
| 6.9967 | 0.176777 | standard |
| 7.0078 | 0.161197 | standard |
| 3.83454 | 1.0 | standard |
| 6.7682 | 0.0884756 | standard |
| 6.77172 | 0.0911445 | standard |
| 6.77811 | 0.0977016 | standard |
| 6.78153 | 0.0967921 | standard |
| 6.94098 | 0.182417 | standard |
| 6.94449 | 0.180851 | standard |
| 6.99729 | 0.175471 | standard |
| 7.00711 | 0.162342 | standard |
| 3.83454 | 1.0 | standard |
| 6.7686 | 0.0894869 | standard |
| 6.77182 | 0.0913371 | standard |
| 6.77791 | 0.0971453 | standard |
| 6.78123 | 0.0961833 | standard |
| 6.94108 | 0.182069 | standard |
| 6.9444 | 0.181306 | standard |
| 6.99759 | 0.175079 | standard |
| 7.00691 | 0.162653 | standard |
| 3.83454 | 1.0 | standard |
| 6.76889 | 0.0894009 | standard |
| 6.77202 | 0.0912764 | standard |
| 6.77781 | 0.097542 | standard |
| 6.78094 | 0.0957994 | standard |
| 6.94118 | 0.182019 | standard |
| 6.9443 | 0.182029 | standard |
| 6.99779 | 0.174862 | standard |
| 7.00661 | 0.163046 | standard |
| 3.83454 | 1.0 | standard |
| 6.77038 | 0.0911354 | standard |
| 6.77272 | 0.0929466 | standard |
| 6.77721 | 0.0964529 | standard |
| 6.77955 | 0.0945538 | standard |
| 6.94157 | 0.182625 | standard |
| 6.94391 | 0.182475 | standard |
| 6.99878 | 0.172127 | standard |
| 7.00562 | 0.165353 | standard |