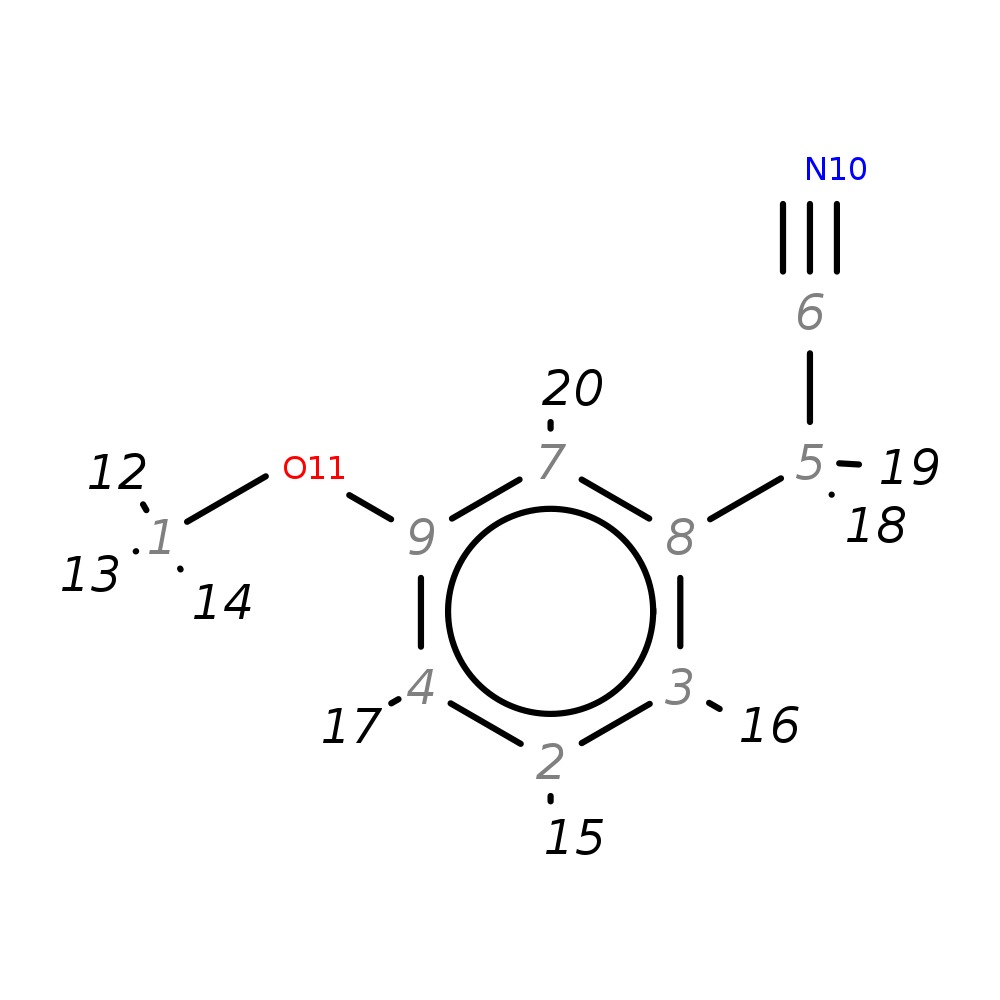
2-(3-Methoxyphenyl)acetonitrile
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03108 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H9NO/c1-11-9-4-2-3-8(7-9)5-6-10/h2-4,7H,5H2,1H3 | |
| Note 1 | 16?17 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(3-Methoxyphenyl)acetonitrile | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|
| 12 | 3.846 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 3.846 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 3.846 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.389 | 6.587 | 7.404 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 6.994 | 0 | 0 | 0 | 2.073 |
| 17 | 0 | 0 | 0 | 0 | 0 | 7.039 | 0 | 0 | 1.323 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 3.922 | -14.0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.922 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.015 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.666717 | standard |
| 6.98644 | 0.0948986 | standard |
| 6.98957 | 0.11397 | standard |
| 6.99736 | 0.0954966 | standard |
| 7.00048 | 0.122561 | standard |
| 7.01459 | 0.195631 | standard |
| 7.0324 | 0.125333 | standard |
| 7.0448 | 0.127124 | standard |
| 7.37743 | 0.0893395 | standard |
| 7.389 | 0.144518 | standard |
| 7.40062 | 0.084863 | standard |
| 3.84623 | 1.0 | standard |
| 3.92089 | 0.71808 | standard |
| 7.03922 | 0.405085 | standard |
| 7.09058 | 0.289643 | standard |
| 7.25294 | 0.152203 | standard |
| 7.37445 | 0.0861184 | standard |
| 7.4172 | 0.0627401 | standard |
| 7.56141 | 0.0235211 | standard |
| 7.59553 | 0.0258341 | standard |
| 3.84585 | 1.0 | standard |
| 3.92178 | 0.691426 | standard |
| 7.02927 | 0.402138 | standard |
| 7.08277 | 0.222333 | standard |
| 7.28835 | 0.114833 | standard |
| 7.36711 | 0.0632051 | standard |
| 7.40212 | 0.0898303 | standard |
| 7.5207 | 0.03413 | standard |
| 3.84578 | 1.00001 | standard |
| 3.92194 | 0.680948 | standard |
| 6.96012 | 0.140614 | standard |
| 7.02362 | 0.411121 | standard |
| 7.07624 | 0.194278 | standard |
| 7.3104 | 0.0971592 | standard |
| 7.39461 | 0.0978901 | standard |
| 7.48439 | 0.040246 | standard |
| 3.84577 | 1.0 | standard |
| 3.92197 | 0.678023 | standard |
| 6.96374 | 0.135586 | standard |
| 7.02149 | 0.417838 | standard |
| 7.0733 | 0.18627 | standard |
| 7.31862 | 0.0918496 | standard |
| 7.3922 | 0.0994195 | standard |
| 7.47254 | 0.0431002 | standard |
| 3.84576 | 1.0 | standard |
| 3.92199 | 0.675934 | standard |
| 6.96664 | 0.13033 | standard |
| 7.0196 | 0.427974 | standard |
| 7.07079 | 0.180328 | standard |
| 7.32539 | 0.0888411 | standard |
| 7.39087 | 0.100486 | standard |
| 7.46315 | 0.046005 | standard |
| 3.84575 | 1.0 | standard |
| 3.92202 | 0.668911 | standard |
| 6.98005 | 0.111921 | standard |
| 7.01263 | 0.440895 | standard |
| 7.05628 | 0.154077 | standard |
| 7.35662 | 0.0803498 | standard |
| 7.39242 | 0.118749 | standard |
| 7.42527 | 0.0603471 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.667652 | standard |
| 6.97924 | 0.088359 | standard |
| 6.98516 | 0.117673 | standard |
| 7.00943 | 0.245684 | standard |
| 7.01348 | 0.24335 | standard |
| 7.02066 | 0.179798 | standard |
| 7.02441 | 0.18261 | standard |
| 7.05017 | 0.138308 | standard |
| 7.36676 | 0.09289 | standard |
| 7.38974 | 0.135281 | standard |
| 7.41237 | 0.0744657 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.667277 | standard |
| 6.98283 | 0.0912103 | standard |
| 6.98741 | 0.115809 | standard |
| 6.99929 | 0.0945678 | standard |
| 7.00381 | 0.151898 | standard |
| 7.01425 | 0.210557 | standard |
| 7.02875 | 0.136658 | standard |
| 7.04744 | 0.133521 | standard |
| 7.37193 | 0.0929133 | standard |
| 7.38921 | 0.141636 | standard |
| 7.40652 | 0.0797866 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.666988 | standard |
| 6.985 | 0.0931695 | standard |
| 6.98877 | 0.11503 | standard |
| 6.99804 | 0.094106 | standard |
| 7.00183 | 0.13108 | standard |
| 7.01446 | 0.200142 | standard |
| 7.03098 | 0.128588 | standard |
| 7.04586 | 0.129948 | standard |
| 7.3752 | 0.0907719 | standard |
| 7.38907 | 0.142809 | standard |
| 7.40295 | 0.0828614 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.666812 | standard |
| 6.98644 | 0.0949272 | standard |
| 6.98957 | 0.114055 | standard |
| 6.99736 | 0.095499 | standard |
| 7.00048 | 0.122534 | standard |
| 7.01459 | 0.195602 | standard |
| 7.0324 | 0.125352 | standard |
| 7.0448 | 0.127102 | standard |
| 7.37743 | 0.0894135 | standard |
| 7.389 | 0.144531 | standard |
| 7.40062 | 0.084856 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.66678 | standard |
| 6.98744 | 0.0955441 | standard |
| 6.99018 | 0.113051 | standard |
| 6.99686 | 0.096722 | standard |
| 6.99954 | 0.11791 | standard |
| 7.01465 | 0.194658 | standard |
| 7.03348 | 0.124656 | standard |
| 7.04401 | 0.12545 | standard |
| 7.37902 | 0.0884024 | standard |
| 7.38894 | 0.143417 | standard |
| 7.39894 | 0.0855557 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.666785 | standard |
| 6.98793 | 0.0964973 | standard |
| 6.99042 | 0.112864 | standard |
| 6.99663 | 0.0967861 | standard |
| 6.99919 | 0.11611 | standard |
| 7.01473 | 0.193712 | standard |
| 7.03386 | 0.123745 | standard |
| 7.04376 | 0.125286 | standard |
| 7.37966 | 0.0884061 | standard |
| 7.38894 | 0.143621 | standard |
| 7.39824 | 0.0862396 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.666499 | standard |
| 6.98826 | 0.0963967 | standard |
| 6.99063 | 0.112181 | standard |
| 6.99643 | 0.0976113 | standard |
| 6.9988 | 0.115029 | standard |
| 7.01472 | 0.193142 | standard |
| 7.03428 | 0.125996 | standard |
| 7.04349 | 0.12445 | standard |
| 7.38021 | 0.0876057 | standard |
| 7.38895 | 0.145132 | standard |
| 7.39765 | 0.0863131 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.666611 | standard |
| 6.98889 | 0.0973499 | standard |
| 6.99098 | 0.112004 | standard |
| 6.9961 | 0.0977774 | standard |
| 6.99826 | 0.112312 | standard |
| 7.01477 | 0.191575 | standard |
| 7.03482 | 0.122999 | standard |
| 7.04307 | 0.124375 | standard |
| 7.38115 | 0.0873081 | standard |
| 7.38893 | 0.146794 | standard |
| 7.39666 | 0.0866502 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.66675 | standard |
| 6.98907 | 0.0973021 | standard |
| 6.99108 | 0.111366 | standard |
| 6.996 | 0.098343 | standard |
| 6.99801 | 0.11204 | standard |
| 7.01476 | 0.191405 | standard |
| 7.035 | 0.121782 | standard |
| 7.03584 | 0.116862 | standard |
| 7.04286 | 0.123109 | standard |
| 7.38154 | 0.0872991 | standard |
| 7.38892 | 0.144545 | standard |
| 7.39626 | 0.0866367 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.66663 | standard |
| 6.98936 | 0.0974342 | standard |
| 6.99128 | 0.111066 | standard |
| 6.99589 | 0.0987366 | standard |
| 6.99782 | 0.111371 | standard |
| 7.01475 | 0.190181 | standard |
| 7.03532 | 0.124153 | standard |
| 7.04262 | 0.120578 | standard |
| 7.0435 | 0.116606 | standard |
| 7.38194 | 0.0873482 | standard |
| 7.38888 | 0.147298 | standard |
| 7.39591 | 0.086846 | standard |
| 3.84574 | 1.0 | standard |
| 3.92202 | 0.666718 | standard |
| 6.99033 | 0.0988235 | standard |
| 6.99179 | 0.109822 | standard |
| 6.99537 | 0.100408 | standard |
| 6.99684 | 0.109005 | standard |
| 7.01484 | 0.190221 | standard |
| 7.03629 | 0.123174 | standard |
| 7.03675 | 0.121174 | standard |
| 7.04188 | 0.11856 | standard |
| 7.04261 | 0.115556 | standard |
| 7.38353 | 0.0871622 | standard |
| 7.38888 | 0.148341 | standard |
| 7.39423 | 0.0871442 | standard |