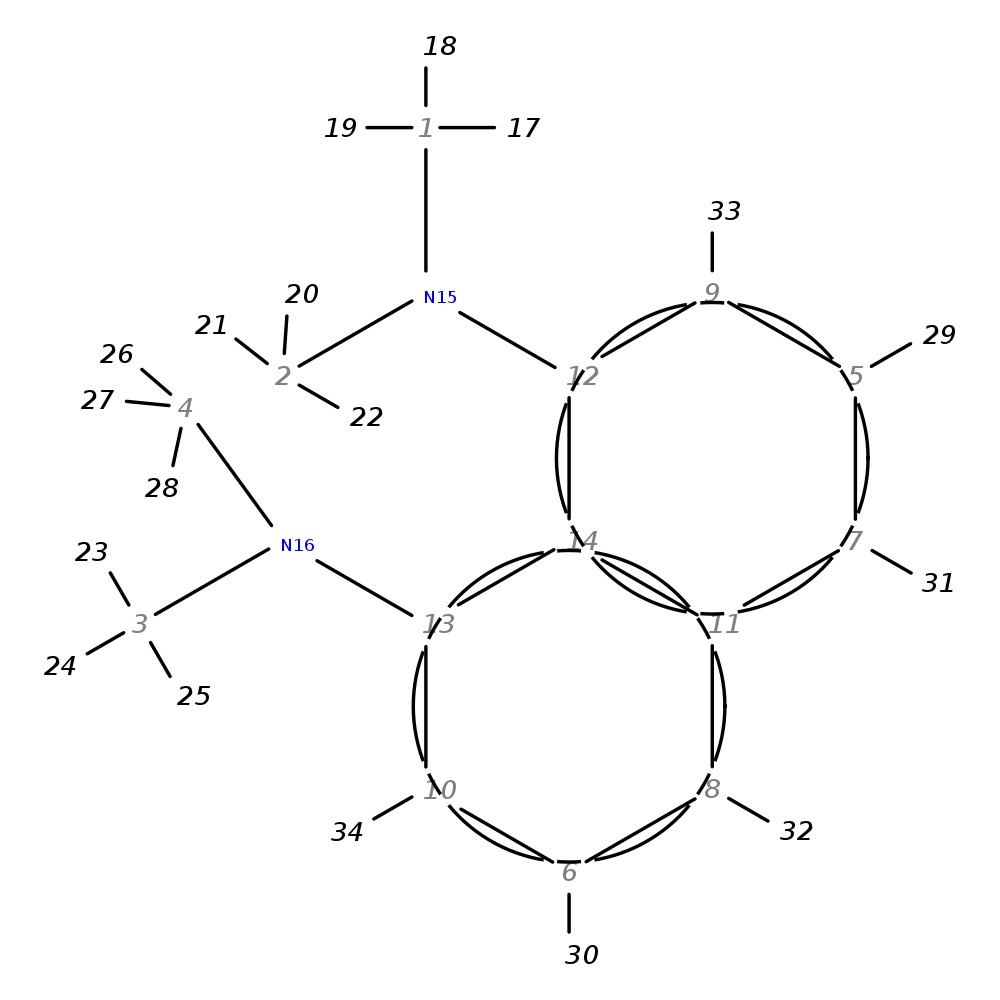
N1,N1,N8,N8-tetramethylnaphthalene-1,8-diamine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02428 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C14H18N2/c1-15(2)12-9-5-7-11-8-6-10-13(14(11)12)16(3)4/h5-10H,1-4H3 | |
| Note 1 | 33,34?31,32 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N1,N1,N8,N8-tetramethylnaphthalene-1,8-diamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 3.154 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 3.154 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 3.154 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 3.154 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 3.154 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 3.154 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 3.154 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.154 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.154 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.154 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.154 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.154 | 0 | 0 | 0 | 0 | 0 | 0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.716 | 0 | 8.155 | 0 | 8.873 | 0 |
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.716 | 0 | 8.155 | 0 | 8.873 |
| 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.931 | 1.5 | 0 | 0 |
| 32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.931 | 0 | 0 |
| 33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.058 | 0 |
| 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.058 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.15432 | 1.0 | standard |
| 7.70137 | 0.032404 | standard |
| 7.71539 | 0.0638694 | standard |
| 7.72945 | 0.0395191 | standard |
| 7.92413 | 0.0748428 | standard |
| 7.93744 | 0.0658596 | standard |
| 8.05102 | 0.0879246 | standard |
| 8.06579 | 0.080823 | standard |
| 3.15432 | 1.0 | standard |
| 7.45181 | 0.0103931 | standard |
| 7.64017 | 0.0304961 | standard |
| 7.84603 | 0.161884 | standard |
| 7.98167 | 0.109537 | standard |
| 8.21505 | 0.0271501 | standard |
| 3.15432 | 1.0 | standard |
| 7.54763 | 0.0144651 | standard |
| 7.67981 | 0.047918 | standard |
| 7.81645 | 0.0944751 | standard |
| 7.88481 | 0.112681 | standard |
| 8.00484 | 0.131864 | standard |
| 8.14666 | 0.044096 | standard |
| 3.15432 | 1.0 | standard |
| 7.59355 | 0.0175411 | standard |
| 7.69494 | 0.0549241 | standard |
| 7.79877 | 0.0727326 | standard |
| 7.89223 | 0.102992 | standard |
| 8.01331 | 0.113348 | standard |
| 8.12108 | 0.055725 | standard |
| 3.15432 | 1.0 | standard |
| 7.60848 | 0.018769 | standard |
| 7.69918 | 0.0569645 | standard |
| 7.79164 | 0.0670682 | standard |
| 7.89532 | 0.099583 | standard |
| 7.98419 | 0.055063 | standard |
| 8.01765 | 0.106963 | standard |
| 8.11332 | 0.0595941 | standard |
| 3.15432 | 1.0 | standard |
| 7.62023 | 0.0199343 | standard |
| 7.70224 | 0.0583482 | standard |
| 7.78571 | 0.0631001 | standard |
| 7.89803 | 0.0966183 | standard |
| 7.97738 | 0.0523611 | standard |
| 8.02095 | 0.104712 | standard |
| 8.10728 | 0.0625221 | standard |
| 3.15432 | 1.0 | standard |
| 7.67054 | 0.0265133 | standard |
| 7.71229 | 0.0630228 | standard |
| 7.75436 | 0.0483403 | standard |
| 7.91247 | 0.0830875 | standard |
| 7.95234 | 0.0581523 | standard |
| 8.0377 | 0.0948611 | standard |
| 8.08167 | 0.0738833 | standard |
| 3.15432 | 1.0 | standard |
| 7.68625 | 0.0291182 | standard |
| 7.71421 | 0.063353 | standard |
| 7.74232 | 0.0435934 | standard |
| 7.91808 | 0.0790392 | standard |
| 7.94467 | 0.0620209 | standard |
| 8.04414 | 0.0916466 | standard |
| 8.07358 | 0.0774954 | standard |
| 3.15432 | 1.0 | standard |
| 7.69392 | 0.0306948 | standard |
| 7.71487 | 0.0634187 | standard |
| 7.73592 | 0.0416002 | standard |
| 7.92102 | 0.0765697 | standard |
| 7.941 | 0.0639772 | standard |
| 8.04754 | 0.0897875 | standard |
| 8.06965 | 0.0791623 | standard |
| 3.15432 | 1.0 | standard |
| 7.69832 | 0.0316073 | standard |
| 7.71518 | 0.0639041 | standard |
| 7.73199 | 0.0404545 | standard |
| 7.92286 | 0.0755088 | standard |
| 7.93878 | 0.0651061 | standard |
| 8.0496 | 0.0886634 | standard |
| 8.06731 | 0.0802052 | standard |
| 3.15432 | 1.0 | standard |
| 7.70137 | 0.032405 | standard |
| 7.71539 | 0.0638664 | standard |
| 7.72945 | 0.0395211 | standard |
| 7.92413 | 0.0748438 | standard |
| 7.93744 | 0.0658596 | standard |
| 8.05102 | 0.0879246 | standard |
| 8.06579 | 0.0808227 | standard |
| 3.15432 | 1.0 | standard |
| 7.70343 | 0.0327187 | standard |
| 7.71542 | 0.0634321 | standard |
| 7.72748 | 0.0387037 | standard |
| 7.925 | 0.0735477 | standard |
| 7.93636 | 0.0667605 | standard |
| 8.05204 | 0.0870801 | standard |
| 8.06469 | 0.0816674 | standard |
| 3.15432 | 1.0 | standard |
| 7.70432 | 0.0330838 | standard |
| 7.71549 | 0.0636321 | standard |
| 7.72669 | 0.0387849 | standard |
| 7.92539 | 0.0738681 | standard |
| 7.93597 | 0.0669838 | standard |
| 8.05244 | 0.0869002 | standard |
| 8.06424 | 0.0819342 | standard |
| 3.15432 | 1.0 | standard |
| 7.705 | 0.0330923 | standard |
| 7.71555 | 0.0638079 | standard |
| 7.72602 | 0.0385238 | standard |
| 7.92568 | 0.0740766 | standard |
| 7.93569 | 0.0667085 | standard |
| 8.05277 | 0.0866123 | standard |
| 8.06384 | 0.0820842 | standard |
| 3.15432 | 1.0 | standard |
| 7.70617 | 0.0333711 | standard |
| 7.71561 | 0.0640841 | standard |
| 7.72493 | 0.0379386 | standard |
| 7.92616 | 0.0726524 | standard |
| 7.9351 | 0.0669508 | standard |
| 8.05339 | 0.086161 | standard |
| 8.06324 | 0.0825032 | standard |
| 3.15432 | 1.0 | standard |
| 7.70677 | 0.0343263 | standard |
| 7.71558 | 0.0632935 | standard |
| 7.72443 | 0.0375863 | standard |
| 7.92646 | 0.072809 | standard |
| 7.9348 | 0.0677816 | standard |
| 8.05363 | 0.0851242 | standard |
| 8.06295 | 0.0836702 | standard |
| 3.15432 | 1.0 | standard |
| 7.70716 | 0.0344126 | standard |
| 7.71558 | 0.0628772 | standard |
| 7.72405 | 0.0372957 | standard |
| 7.92666 | 0.0714925 | standard |
| 7.93462 | 0.0680439 | standard |
| 8.05382 | 0.0849912 | standard |
| 8.06275 | 0.0837652 | standard |
| 3.15432 | 1.0 | standard |
| 7.70923 | 0.0347779 | standard |
| 7.71564 | 0.0641059 | standard |
| 7.72219 | 0.0363611 | standard |
| 7.92753 | 0.07171 | standard |
| 7.93365 | 0.0694824 | standard |
| 8.05482 | 0.0847192 | standard |
| 8.06166 | 0.0841002 | standard |