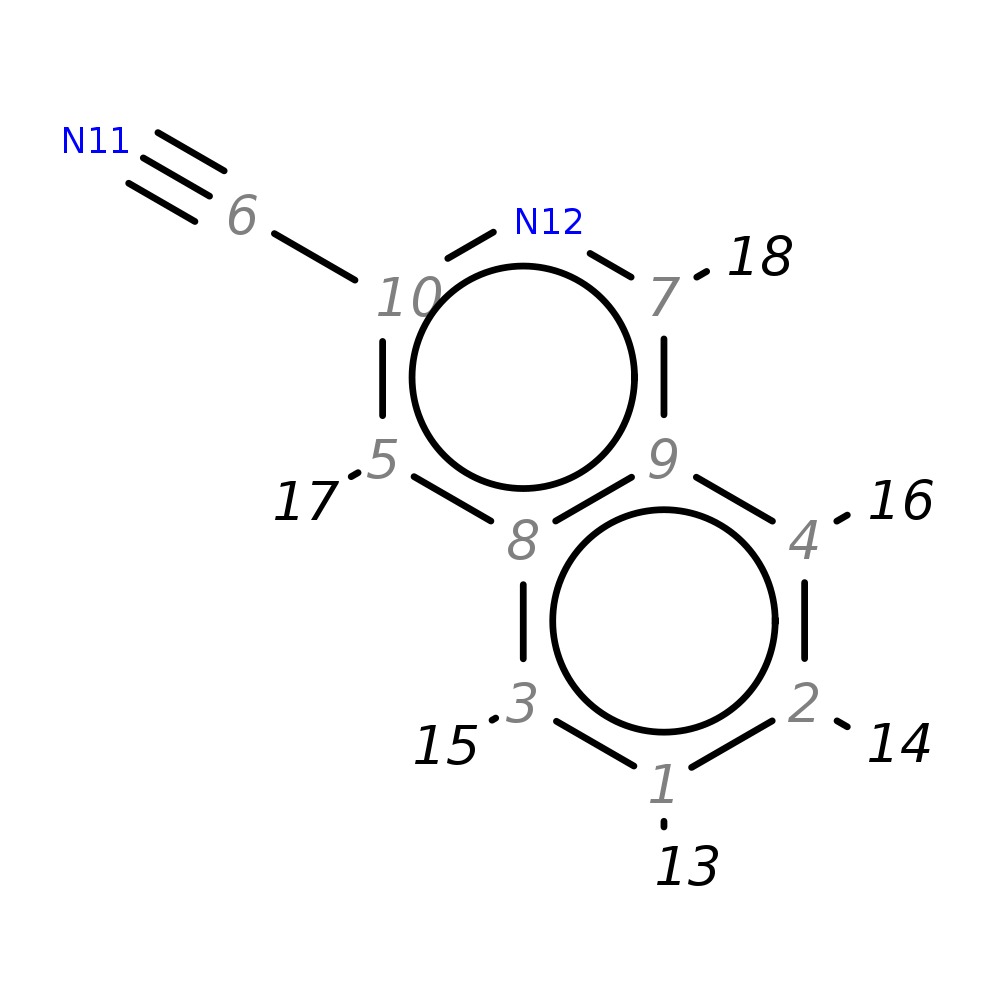
3-Isoquinolinecarbonitrile
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01480 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H6N2/c11-6-10-5-8-3-1-2-4-9(8)7-12-10/h1-5,7H | |
| Note 1 | Aliphatic region not clean | |
| Note 2 | 17?18 | |
| Note 3 | 15?16 | |
| Note 4 | 13?14 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Isoquinolinecarbonitrile | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|
| 13 | 7.935 | 7.206 | 8.746 | 0 | 0 | 0 |
| 14 | 0 | 7.986 | 0 | 8.573 | 0 | 0 |
| 15 | 0 | 0 | 8.104 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 8.244 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 8.44 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 8.487 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 7.92093 | 0.189311 | standard |
| 7.93315 | 0.35314 | standard |
| 7.94724 | 0.348401 | standard |
| 7.97354 | 0.312378 | standard |
| 7.98741 | 0.396881 | standard |
| 7.99956 | 0.222096 | standard |
| 8.09739 | 0.547248 | standard |
| 8.11172 | 0.464119 | standard |
| 8.2372 | 0.530029 | standard |
| 8.25128 | 0.475016 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999965 | standard |
| 7.81662 | 0.100152 | standard |
| 7.87831 | 0.139735 | standard |
| 8.03777 | 1.0 | standard |
| 8.1677 | 0.427439 | standard |
| 8.26831 | 0.14527 | standard |
| 8.29866 | 0.140908 | standard |
| 8.33626 | 0.155732 | standard |
| 8.44266 | 0.828837 | standard |
| 8.48446 | 0.816771 | standard |
| 7.77744 | 0.0404631 | standard |
| 7.87174 | 0.202069 | standard |
| 7.92501 | 0.27794 | standard |
| 7.95872 | 0.433372 | standard |
| 8.00626 | 0.796838 | standard |
| 8.01763 | 0.794774 | standard |
| 8.05435 | 0.769333 | standard |
| 8.13416 | 0.336479 | standard |
| 8.19094 | 0.447595 | standard |
| 8.26291 | 0.191915 | standard |
| 8.31007 | 0.155901 | standard |
| 8.33614 | 0.176988 | standard |
| 8.44094 | 1.0 | standard |
| 8.48665 | 0.995334 | standard |
| 7.8154 | 0.048221 | standard |
| 7.89888 | 0.255076 | standard |
| 7.95552 | 0.417554 | standard |
| 7.99403 | 0.689287 | standard |
| 8.0113 | 0.636075 | standard |
| 8.06575 | 0.624502 | standard |
| 8.12016 | 0.245409 | standard |
| 8.20081 | 0.441104 | standard |
| 8.2559 | 0.214426 | standard |
| 8.30821 | 0.201451 | standard |
| 8.35257 | 0.066175 | standard |
| 8.44056 | 1.0 | standard |
| 8.48706 | 0.997817 | standard |
| 7.82894 | 0.0526172 | standard |
| 7.90899 | 0.268789 | standard |
| 7.95442 | 0.444016 | standard |
| 7.99129 | 0.651775 | standard |
| 8.00683 | 0.57027 | standard |
| 8.06729 | 0.629969 | standard |
| 8.11961 | 0.213996 | standard |
| 8.16848 | 0.253762 | standard |
| 8.20543 | 0.429453 | standard |
| 8.25209 | 0.201015 | standard |
| 8.29966 | 0.216073 | standard |
| 8.33908 | 0.057752 | standard |
| 8.44048 | 1.00001 | standard |
| 8.48713 | 0.998332 | standard |
| 7.83971 | 0.0569993 | standard |
| 7.91714 | 0.280182 | standard |
| 7.95389 | 0.478796 | standard |
| 7.98885 | 0.580041 | standard |
| 8.00335 | 0.540095 | standard |
| 8.06976 | 0.557068 | standard |
| 8.10893 | 0.181034 | standard |
| 8.12117 | 0.175952 | standard |
| 8.16232 | 0.22867 | standard |
| 8.20866 | 0.421805 | standard |
| 8.24638 | 0.181693 | standard |
| 8.29302 | 0.231382 | standard |
| 8.32955 | 0.053082 | standard |
| 8.44044 | 1.0 | standard |
| 8.48718 | 0.998668 | standard |
| 7.88887 | 0.103703 | standard |
| 7.92382 | 0.313965 | standard |
| 7.95872 | 0.627281 | standard |
| 7.99249 | 0.426797 | standard |
| 8.02686 | 0.1813 | standard |
| 8.0574 | 0.0577361 | standard |
| 8.08596 | 0.475887 | standard |
| 8.12814 | 0.297416 | standard |
| 8.15633 | 0.0291828 | standard |
| 8.1967 | 0.0445757 | standard |
| 8.22527 | 0.447226 | standard |
| 8.26626 | 0.331573 | standard |
| 8.29443 | 0.0274601 | standard |
| 8.44036 | 1.0 | standard |
| 8.48726 | 0.999684 | standard |
| 7.90533 | 0.137917 | standard |
| 7.92938 | 0.349766 | standard |
| 7.9571 | 0.476915 | standard |
| 7.96145 | 0.430624 | standard |
| 7.98963 | 0.414124 | standard |
| 8.01348 | 0.192409 | standard |
| 8.06084 | 0.0192775 | standard |
| 8.09157 | 0.524501 | standard |
| 8.1194 | 0.38297 | standard |
| 8.14956 | 0.0117981 | standard |
| 8.2006 | 0.0161122 | standard |
| 8.23119 | 0.503162 | standard |
| 8.2584 | 0.411455 | standard |
| 8.28871 | 0.011314 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.99986 | standard |
| 7.91326 | 0.161595 | standard |
| 7.93145 | 0.353428 | standard |
| 7.95223 | 0.394252 | standard |
| 7.96798 | 0.340688 | standard |
| 7.98844 | 0.405688 | standard |
| 8.00652 | 0.204626 | standard |
| 8.0607 | 0.00825833 | standard |
| 8.09435 | 0.548537 | standard |
| 8.11553 | 0.427589 | standard |
| 8.23411 | 0.525961 | standard |
| 8.25484 | 0.450682 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999922 | standard |
| 7.91791 | 0.177698 | standard |
| 7.93254 | 0.353322 | standard |
| 7.94932 | 0.367342 | standard |
| 7.97127 | 0.324852 | standard |
| 7.9878 | 0.40153 | standard |
| 8.00237 | 0.213785 | standard |
| 8.0961 | 0.550797 | standard |
| 8.11323 | 0.449518 | standard |
| 8.23593 | 0.529343 | standard |
| 8.25269 | 0.466562 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.99995 | standard |
| 7.92093 | 0.18931 | standard |
| 7.93315 | 0.353142 | standard |
| 7.94724 | 0.348403 | standard |
| 7.97354 | 0.31238 | standard |
| 7.98741 | 0.396884 | standard |
| 7.99956 | 0.222096 | standard |
| 8.09739 | 0.54725 | standard |
| 8.11172 | 0.464119 | standard |
| 8.2372 | 0.530029 | standard |
| 8.25128 | 0.475016 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999965 | standard |
| 7.92306 | 0.19826 | standard |
| 7.93357 | 0.354944 | standard |
| 7.9457 | 0.33475 | standard |
| 7.97522 | 0.304829 | standard |
| 7.98713 | 0.390204 | standard |
| 7.99763 | 0.226714 | standard |
| 8.09831 | 0.543504 | standard |
| 8.11065 | 0.469897 | standard |
| 8.23809 | 0.526499 | standard |
| 8.25023 | 0.48053 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999974 | standard |
| 7.92395 | 0.201899 | standard |
| 7.93365 | 0.350244 | standard |
| 7.93497 | 0.315901 | standard |
| 7.94506 | 0.330102 | standard |
| 7.97592 | 0.301138 | standard |
| 7.98704 | 0.38717 | standard |
| 7.99683 | 0.228465 | standard |
| 8.09863 | 0.53955 | standard |
| 8.11021 | 0.47358 | standard |
| 8.23849 | 0.524972 | standard |
| 8.24984 | 0.482001 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999978 | standard |
| 7.92464 | 0.204889 | standard |
| 7.93385 | 0.35474 | standard |
| 7.94447 | 0.325149 | standard |
| 7.97651 | 0.297928 | standard |
| 7.98694 | 0.388212 | standard |
| 7.99614 | 0.230034 | standard |
| 8.099 | 0.538569 | standard |
| 8.10981 | 0.475456 | standard |
| 8.23886 | 0.525026 | standard |
| 8.24947 | 0.48459 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.99998 | standard |
| 7.92583 | 0.210127 | standard |
| 7.93403 | 0.351085 | standard |
| 7.93515 | 0.323355 | standard |
| 7.94347 | 0.316973 | standard |
| 7.9775 | 0.292711 | standard |
| 7.98686 | 0.379825 | standard |
| 7.995 | 0.233169 | standard |
| 8.09955 | 0.53565 | standard |
| 8.10922 | 0.478382 | standard |
| 8.2394 | 0.523873 | standard |
| 8.24887 | 0.486807 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999984 | standard |
| 7.92637 | 0.212962 | standard |
| 7.93406 | 0.348801 | standard |
| 7.9352 | 0.32085 | standard |
| 7.94308 | 0.313488 | standard |
| 7.978 | 0.290755 | standard |
| 7.98676 | 0.38036 | standard |
| 7.9945 | 0.234337 | standard |
| 8.0998 | 0.532953 | standard |
| 8.10892 | 0.479692 | standard |
| 8.23963 | 0.522177 | standard |
| 8.2486 | 0.488008 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999986 | standard |
| 7.92682 | 0.214495 | standard |
| 7.93416 | 0.350099 | standard |
| 7.93521 | 0.324099 | standard |
| 7.94273 | 0.311136 | standard |
| 7.97835 | 0.289489 | standard |
| 7.98672 | 0.378194 | standard |
| 7.99406 | 0.234882 | standard |
| 8.1 | 0.531343 | standard |
| 8.10872 | 0.480716 | standard |
| 8.23985 | 0.52099 | standard |
| 8.24838 | 0.488531 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999987 | standard |
| 7.9288 | 0.223579 | standard |
| 7.93442 | 0.344758 | standard |
| 7.93532 | 0.323354 | standard |
| 7.94105 | 0.29831 | standard |
| 7.98008 | 0.280547 | standard |
| 7.98654 | 0.372451 | standard |
| 7.99222 | 0.240318 | standard |
| 8.10094 | 0.526479 | standard |
| 8.10768 | 0.487336 | standard |
| 8.24079 | 0.518918 | standard |
| 8.24733 | 0.493813 | standard |
| 8.44035 | 1.0 | standard |
| 8.48727 | 0.999993 | standard |