3-Methoxyphenethyl alcohol
Simulation outputs:
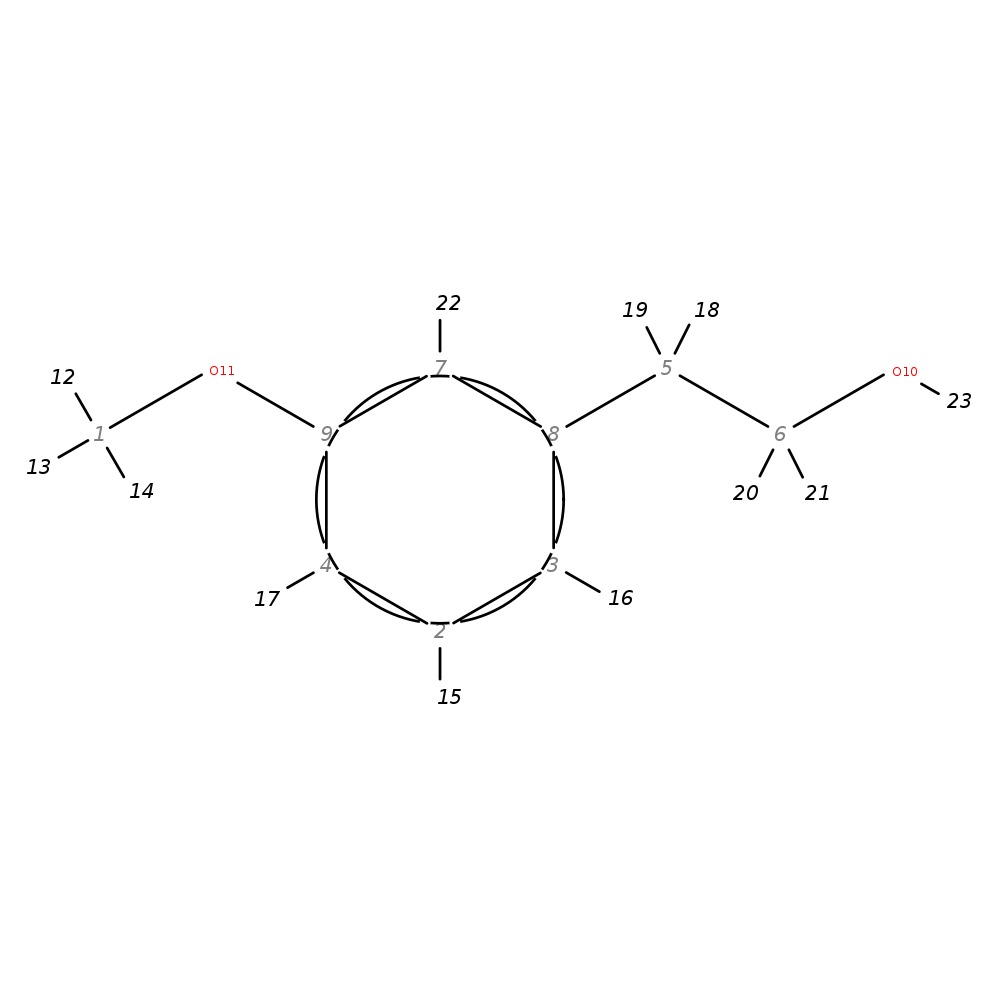
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01147 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H12O2/c1-11-9-4-2-3-8(7-9)5-6-10/h2-4,7,10H,5-6H2,1H3 | |
| Note 1 | 19,18?20,21 | |
| Note 2 | 17?16 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Methoxyphenethyl alcohol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 3.83 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 3.83 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 3.83 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.319 | 8.249 | 8.249 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 6.942 | 0 | 0 | 0 | 0 | 0 | 1.627 |
| 17 | 0 | 0 | 0 | 0 | 0 | 6.893 | 0 | 0 | 0 | 0 | 2.395 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 2.843 | -14.0 | 7.035 | 7.035 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.843 | 7.035 | 7.035 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.83 | -14.0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.83 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.919 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.83103 | 0.130079 | standard |
| 2.84272 | 0.253862 | standard |
| 2.85447 | 0.129709 | standard |
| 3.81831 | 0.144922 | standard |
| 3.8299 | 1.00021 | standard |
| 3.84169 | 0.14459 | standard |
| 6.88395 | 0.0655181 | standard |
| 6.88773 | 0.0790428 | standard |
| 6.8976 | 0.0678237 | standard |
| 6.90137 | 0.0890285 | standard |
| 6.91868 | 0.138496 | standard |
| 6.92193 | 0.0950538 | standard |
| 6.93447 | 0.089682 | standard |
| 6.9365 | 0.0766013 | standard |
| 6.9482 | 0.0905232 | standard |
| 6.95031 | 0.0825059 | standard |
| 7.30522 | 0.0687254 | standard |
| 7.31888 | 0.126054 | standard |
| 7.33253 | 0.0601149 | standard |
| 2.64683 | 0.0696991 | standard |
| 2.66672 | 0.073309 | standard |
| 2.83657 | 0.201564 | standard |
| 3.00142 | 0.159874 | standard |
| 3.67371 | 0.178971 | standard |
| 3.83057 | 1.0 | standard |
| 4.00439 | 0.0890093 | standard |
| 6.78087 | 0.074683 | standard |
| 6.87504 | 0.157922 | standard |
| 6.95052 | 0.331982 | standard |
| 6.99322 | 0.252771 | standard |
| 7.16333 | 0.135151 | standard |
| 7.30373 | 0.0685853 | standard |
| 7.36641 | 0.0434581 | standard |
| 7.45628 | 0.0187981 | standard |
| 7.52392 | 0.0167241 | standard |
| 7.56538 | 0.0192231 | standard |
| 2.72043 | 0.0919161 | standard |
| 2.83722 | 0.224324 | standard |
| 2.95226 | 0.152517 | standard |
| 3.72113 | 0.170191 | standard |
| 3.83053 | 1.0 | standard |
| 3.95098 | 0.106215 | standard |
| 6.79331 | 0.0469521 | standard |
| 6.83024 | 0.093142 | standard |
| 6.89022 | 0.16533 | standard |
| 6.93706 | 0.309298 | standard |
| 6.98679 | 0.180074 | standard |
| 7.20246 | 0.0949344 | standard |
| 7.29778 | 0.0485112 | standard |
| 7.34082 | 0.0655871 | standard |
| 7.41501 | 0.0183021 | standard |
| 7.47591 | 0.0240381 | standard |
| 2.75154 | 0.102773 | standard |
| 2.83914 | 0.23693 | standard |
| 2.92644 | 0.148324 | standard |
| 3.74664 | 0.165191 | standard |
| 3.83043 | 1.00001 | standard |
| 3.92068 | 0.117221 | standard |
| 6.82019 | 0.0509554 | standard |
| 6.84777 | 0.0915908 | standard |
| 6.89839 | 0.191494 | standard |
| 6.93035 | 0.306806 | standard |
| 6.98121 | 0.145892 | standard |
| 7.22745 | 0.0765506 | standard |
| 7.33209 | 0.0717173 | standard |
| 7.39696 | 0.0156521 | standard |
| 7.43277 | 0.0279533 | standard |
| 2.76197 | 0.106246 | standard |
| 2.83983 | 0.240615 | standard |
| 2.91756 | 0.146392 | standard |
| 3.75541 | 0.163273 | standard |
| 3.83037 | 1.0 | standard |
| 3.91043 | 0.120464 | standard |
| 6.82912 | 0.0524293 | standard |
| 6.85319 | 0.0898321 | standard |
| 6.90219 | 0.207934 | standard |
| 6.92794 | 0.308439 | standard |
| 6.97867 | 0.137502 | standard |
| 7.23669 | 0.0704883 | standard |
| 7.28478 | 0.0252161 | standard |
| 7.33014 | 0.072396 | standard |
| 7.41865 | 0.029714 | standard |
| 2.77025 | 0.109358 | standard |
| 2.84036 | 0.24335 | standard |
| 2.91042 | 0.145067 | standard |
| 3.76251 | 0.161734 | standard |
| 3.83031 | 1.00001 | standard |
| 3.90216 | 0.123362 | standard |
| 6.83634 | 0.0536731 | standard |
| 6.85736 | 0.0881023 | standard |
| 6.90581 | 0.226023 | standard |
| 6.92593 | 0.310616 | standard |
| 6.97636 | 0.131396 | standard |
| 7.24437 | 0.066927 | standard |
| 7.28141 | 0.0226192 | standard |
| 7.32912 | 0.0726102 | standard |
| 7.40754 | 0.031695 | standard |
| 2.80706 | 0.121822 | standard |
| 2.84218 | 0.2514 | standard |
| 2.87731 | 0.137572 | standard |
| 3.79548 | 0.153397 | standard |
| 3.83002 | 1.00009 | standard |
| 3.86559 | 0.13621 | standard |
| 6.87174 | 0.0694842 | standard |
| 6.87604 | 0.0706597 | standard |
| 6.91632 | 0.347343 | standard |
| 6.96054 | 0.107729 | standard |
| 7.28021 | 0.0569055 | standard |
| 7.31907 | 0.0899984 | standard |
| 7.36205 | 0.0447123 | standard |
| 2.81909 | 0.12684 | standard |
| 2.84252 | 0.252716 | standard |
| 2.86596 | 0.132207 | standard |
| 3.80685 | 0.147563 | standard |
| 3.82995 | 1.00007 | standard |
| 3.85355 | 0.14166 | standard |
| 6.87513 | 0.0617598 | standard |
| 6.88182 | 0.080774 | standard |
| 6.90992 | 0.159793 | standard |
| 6.91234 | 0.15728 | standard |
| 6.91798 | 0.16946 | standard |
| 6.92587 | 0.185406 | standard |
| 6.95393 | 0.0990128 | standard |
| 7.29275 | 0.0682465 | standard |
| 7.31966 | 0.115087 | standard |
| 7.34628 | 0.051458 | standard |
| 2.82508 | 0.128964 | standard |
| 2.84262 | 0.253258 | standard |
| 2.86026 | 0.130744 | standard |
| 3.81253 | 0.145969 | standard |
| 3.82992 | 1.00028 | standard |
| 3.84762 | 0.143446 | standard |
| 6.8794 | 0.0631082 | standard |
| 6.88493 | 0.0811597 | standard |
| 6.89976 | 0.0645022 | standard |
| 6.90534 | 0.10745 | standard |
| 6.9184 | 0.147453 | standard |
| 6.92373 | 0.106258 | standard |
| 6.93026 | 0.107602 | standard |
| 6.95097 | 0.0942413 | standard |
| 6.95401 | 0.0843162 | standard |
| 7.29877 | 0.0699569 | standard |
| 7.3192 | 0.123964 | standard |
| 7.33947 | 0.0572646 | standard |
| 2.8287 | 0.129707 | standard |
| 2.84267 | 0.253198 | standard |
| 2.85674 | 0.13052 | standard |
| 3.81604 | 0.145838 | standard |
| 3.8299 | 1.00022 | standard |
| 3.84401 | 0.144296 | standard |
| 6.88216 | 0.0648169 | standard |
| 6.88657 | 0.0799513 | standard |
| 6.89845 | 0.0664472 | standard |
| 6.90296 | 0.0947097 | standard |
| 6.91856 | 0.141422 | standard |
| 6.92261 | 0.0971047 | standard |
| 6.93282 | 0.0938481 | standard |
| 6.94932 | 0.0919196 | standard |
| 6.95186 | 0.0826911 | standard |
| 7.30259 | 0.0695207 | standard |
| 7.31895 | 0.125517 | standard |
| 7.33531 | 0.059415 | standard |
| 2.83103 | 0.130133 | standard |
| 2.84272 | 0.253221 | standard |
| 2.85447 | 0.129763 | standard |
| 3.81831 | 0.144989 | standard |
| 3.8299 | 1.00021 | standard |
| 3.84169 | 0.144658 | standard |
| 6.88395 | 0.0655041 | standard |
| 6.88773 | 0.0790258 | standard |
| 6.8976 | 0.0678097 | standard |
| 6.90137 | 0.0890095 | standard |
| 6.91868 | 0.138467 | standard |
| 6.92193 | 0.0950348 | standard |
| 6.93447 | 0.089663 | standard |
| 6.9365 | 0.0765853 | standard |
| 6.9482 | 0.0905042 | standard |
| 6.95031 | 0.0824889 | standard |
| 7.30522 | 0.0687104 | standard |
| 7.31888 | 0.126027 | standard |
| 7.33253 | 0.0601009 | standard |
| 2.83271 | 0.129966 | standard |
| 2.84272 | 0.253212 | standard |
| 2.85283 | 0.12987 | standard |
| 3.81994 | 0.145062 | standard |
| 3.82989 | 1.00007 | standard |
| 3.84001 | 0.144391 | standard |
| 6.88522 | 0.0661959 | standard |
| 6.88848 | 0.0782661 | standard |
| 6.89692 | 0.0684012 | standard |
| 6.90023 | 0.0854446 | standard |
| 6.91873 | 0.136999 | standard |
| 6.92159 | 0.0927676 | standard |
| 6.93566 | 0.0875756 | standard |
| 6.93742 | 0.0773574 | standard |
| 6.94737 | 0.0889938 | standard |
| 6.94925 | 0.0816674 | standard |
| 7.3071 | 0.0679968 | standard |
| 7.3188 | 0.126113 | standard |
| 7.3305 | 0.0607409 | standard |
| 2.83341 | 0.129941 | standard |
| 2.84275 | 0.253111 | standard |
| 2.85214 | 0.129889 | standard |
| 3.82063 | 0.145164 | standard |
| 3.82989 | 1.00008 | standard |
| 3.83932 | 0.14444 | standard |
| 6.88576 | 0.0665964 | standard |
| 6.88878 | 0.0781046 | standard |
| 6.89671 | 0.069195 | standard |
| 6.89973 | 0.08456 | standard |
| 6.91877 | 0.13609 | standard |
| 6.92139 | 0.0932095 | standard |
| 6.9361 | 0.0867651 | standard |
| 6.93776 | 0.0776177 | standard |
| 6.94706 | 0.0884595 | standard |
| 6.94883 | 0.0815437 | standard |
| 7.3078 | 0.0676958 | standard |
| 7.3188 | 0.12608 | standard |
| 7.32971 | 0.0609511 | standard |
| 2.834 | 0.129882 | standard |
| 2.84277 | 0.253683 | standard |
| 2.85154 | 0.129882 | standard |
| 3.82123 | 0.145184 | standard |
| 3.82989 | 1.00009 | standard |
| 3.83872 | 0.14441 | standard |
| 6.8862 | 0.0669969 | standard |
| 6.88903 | 0.0779796 | standard |
| 6.89651 | 0.0695387 | standard |
| 6.89934 | 0.0836028 | standard |
| 6.91884 | 0.13638 | standard |
| 6.92119 | 0.0937174 | standard |
| 6.93649 | 0.08581 | standard |
| 6.93811 | 0.0773272 | standard |
| 6.94675 | 0.0876488 | standard |
| 6.94846 | 0.0809284 | standard |
| 7.30849 | 0.06749 | standard |
| 7.31875 | 0.126286 | standard |
| 7.32901 | 0.0611321 | standard |
| 2.83499 | 0.129889 | standard |
| 2.84277 | 0.252853 | standard |
| 2.8506 | 0.130079 | standard |
| 3.82216 | 0.145322 | standard |
| 3.82989 | 1.00012 | standard |
| 3.83773 | 0.144451 | standard |
| 6.88698 | 0.0676825 | standard |
| 6.88948 | 0.0776011 | standard |
| 6.89609 | 0.0698923 | standard |
| 6.89865 | 0.0819035 | standard |
| 6.91888 | 0.135559 | standard |
| 6.92089 | 0.0941494 | standard |
| 6.93717 | 0.0845844 | standard |
| 6.93866 | 0.07734 | standard |
| 6.94635 | 0.0879862 | standard |
| 6.94777 | 0.0820975 | standard |
| 7.30958 | 0.0670541 | standard |
| 7.31875 | 0.126256 | standard |
| 7.32787 | 0.0615389 | standard |
| 2.83539 | 0.130005 | standard |
| 2.84277 | 0.254217 | standard |
| 2.8502 | 0.130052 | standard |
| 3.82256 | 0.145232 | standard |
| 3.82989 | 1.00013 | standard |
| 3.83734 | 0.144469 | standard |
| 6.88727 | 0.0680642 | standard |
| 6.88963 | 0.0777184 | standard |
| 6.89589 | 0.0699061 | standard |
| 6.89836 | 0.0811352 | standard |
| 6.91886 | 0.135909 | standard |
| 6.92086 | 0.093079 | standard |
| 6.9374 | 0.0838343 | standard |
| 6.93889 | 0.0767768 | standard |
| 6.94615 | 0.0878979 | standard |
| 6.94748 | 0.0822628 | standard |
| 7.31008 | 0.0669642 | standard |
| 7.31873 | 0.12618 | standard |
| 7.32738 | 0.0617359 | standard |
| 2.83574 | 0.130181 | standard |
| 2.84277 | 0.253539 | standard |
| 2.84981 | 0.130181 | standard |
| 3.82291 | 0.144917 | standard |
| 3.82989 | 1.00014 | standard |
| 3.83694 | 0.144505 | standard |
| 6.88756 | 0.0694061 | standard |
| 6.88983 | 0.0788119 | standard |
| 6.89579 | 0.0693693 | standard |
| 6.89806 | 0.0796272 | standard |
| 6.91886 | 0.134673 | standard |
| 6.92068 | 0.0945752 | standard |
| 6.93772 | 0.0852829 | standard |
| 6.93896 | 0.0791324 | standard |
| 6.94595 | 0.0879178 | standard |
| 6.94719 | 0.0825637 | standard |
| 7.31048 | 0.0655438 | standard |
| 7.31873 | 0.126143 | standard |
| 7.32693 | 0.0628589 | standard |
| 2.83742 | 0.130345 | standard |
| 2.84277 | 0.25397 | standard |
| 2.84822 | 0.130166 | standard |
| 3.82454 | 0.145437 | standard |
| 3.82989 | 1.00023 | standard |
| 3.83531 | 0.144897 | standard |
| 6.88884 | 0.0706629 | standard |
| 6.89053 | 0.0783048 | standard |
| 6.89513 | 0.07037 | standard |
| 6.89687 | 0.0778667 | standard |
| 6.91769 | 0.0967804 | standard |
| 6.91894 | 0.133702 | standard |
| 6.92028 | 0.0940855 | standard |
| 6.93889 | 0.0865679 | standard |
| 6.9398 | 0.0822787 | standard |
| 6.94512 | 0.0841337 | standard |
| 6.94618 | 0.0795061 | standard |
| 7.31231 | 0.0643779 | standard |
| 7.31865 | 0.126491 | standard |
| 7.325 | 0.0643779 | standard |