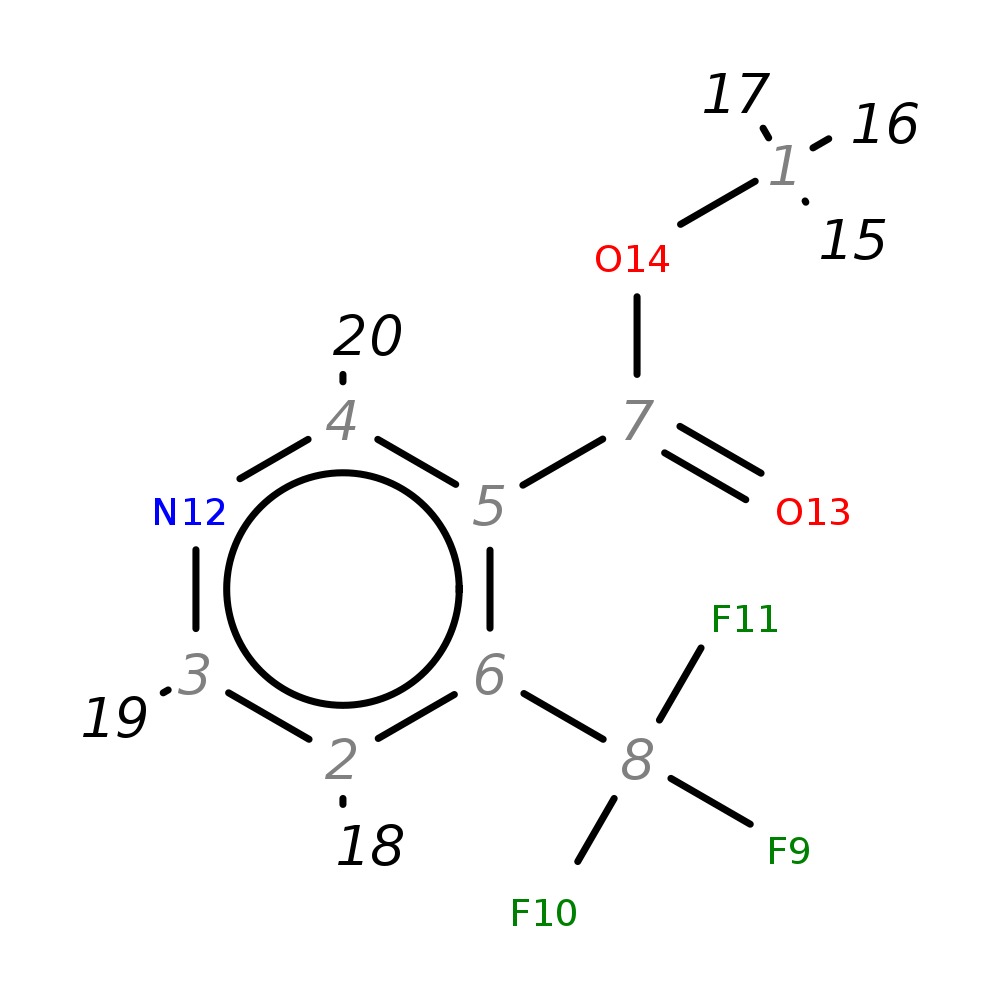
Methyl 4-(trifluoromethyl)nicotinate
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02024 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H6F3NO2/c1-14-7(13)5-4-12-3-2-6(5)8(9,10)11/h2-4H,1H3 | |
| Note 1 | 18?19 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Methyl 4-(trifluoromethyl)nicotinate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|
| 15 | 4.005 | -14.0 | -14.0 | 0 | 0 | 0 |
| 16 | 0 | 4.005 | -14.0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 4.005 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.908 | 5.547 | 0 |
| 19 | 0 | 0 | 0 | 0 | 8.93 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 9.075 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.00484 | 1.0 | standard |
| 7.90307 | 0.171831 | standard |
| 7.91237 | 0.171751 | standard |
| 8.92558 | 0.171899 | standard |
| 8.93479 | 0.171986 | standard |
| 9.07475 | 0.333285 | standard |
| 4.00484 | 1.0 | standard |
| 7.83392 | 0.150508 | standard |
| 7.97229 | 0.193924 | standard |
| 8.86565 | 0.198454 | standard |
| 9.00616 | 0.187534 | standard |
| 9.07434 | 0.352151 | standard |
| 4.00484 | 1.0 | standard |
| 7.85953 | 0.157499 | standard |
| 7.95179 | 0.186101 | standard |
| 8.8861 | 0.189067 | standard |
| 8.97851 | 0.166343 | standard |
| 9.07471 | 0.337868 | standard |
| 4.00484 | 1.0 | standard |
| 7.872 | 0.160801 | standard |
| 7.94119 | 0.182696 | standard |
| 8.89668 | 0.184094 | standard |
| 8.96591 | 0.16525 | standard |
| 9.07474 | 0.335755 | standard |
| 4.00484 | 1.0 | standard |
| 7.87605 | 0.162253 | standard |
| 7.93752 | 0.181558 | standard |
| 8.90034 | 0.183015 | standard |
| 8.96182 | 0.165363 | standard |
| 9.07474 | 0.335419 | standard |
| 4.00484 | 1.0 | standard |
| 7.87932 | 0.163216 | standard |
| 7.93465 | 0.180339 | standard |
| 8.90321 | 0.181661 | standard |
| 8.95855 | 0.165567 | standard |
| 9.07474 | 0.334954 | standard |
| 4.00484 | 1.0 | standard |
| 7.89367 | 0.170086 | standard |
| 7.92138 | 0.173222 | standard |
| 8.91647 | 0.173552 | standard |
| 8.94419 | 0.170567 | standard |
| 9.07475 | 0.333099 | standard |
| 4.00484 | 1.0 | standard |
| 7.89842 | 0.171939 | standard |
| 7.91683 | 0.171833 | standard |
| 8.92103 | 0.172083 | standard |
| 8.93944 | 0.172041 | standard |
| 9.07475 | 0.333399 | standard |
| 4.00484 | 1.0 | standard |
| 7.90079 | 0.171817 | standard |
| 7.91465 | 0.171896 | standard |
| 8.9233 | 0.172058 | standard |
| 8.93706 | 0.171998 | standard |
| 9.07475 | 0.332571 | standard |
| 4.00484 | 1.0 | standard |
| 7.90218 | 0.171901 | standard |
| 7.91327 | 0.171749 | standard |
| 8.92459 | 0.171959 | standard |
| 8.93568 | 0.171816 | standard |
| 9.07475 | 0.33324 | standard |
| 4.00484 | 1.0 | standard |
| 7.90307 | 0.171813 | standard |
| 7.91237 | 0.171914 | standard |
| 8.92558 | 0.172064 | standard |
| 8.93479 | 0.171968 | standard |
| 9.07475 | 0.333424 | standard |
| 4.00484 | 1.0 | standard |
| 7.90376 | 0.171896 | standard |
| 7.91168 | 0.171587 | standard |
| 8.92618 | 0.171924 | standard |
| 8.93409 | 0.171623 | standard |
| 9.07475 | 0.333062 | standard |
| 4.00484 | 1.0 | standard |
| 7.90406 | 0.171997 | standard |
| 7.91138 | 0.171987 | standard |
| 8.92647 | 0.172023 | standard |
| 8.9338 | 0.172016 | standard |
| 9.07475 | 0.333352 | standard |
| 4.00484 | 1.0 | standard |
| 7.90426 | 0.171884 | standard |
| 7.91119 | 0.171251 | standard |
| 8.92667 | 0.171541 | standard |
| 8.9336 | 0.171644 | standard |
| 9.07475 | 0.332598 | standard |
| 4.00484 | 1.0 | standard |
| 7.90465 | 0.171937 | standard |
| 7.91079 | 0.171789 | standard |
| 8.92707 | 0.171938 | standard |
| 8.9332 | 0.171826 | standard |
| 9.07475 | 0.332778 | standard |
| 4.00484 | 1.0 | standard |
| 7.90485 | 0.172185 | standard |
| 7.91059 | 0.172183 | standard |
| 8.92726 | 0.172023 | standard |
| 8.9331 | 0.172026 | standard |
| 9.07475 | 0.333567 | standard |
| 4.00484 | 1.0 | standard |
| 7.90495 | 0.171897 | standard |
| 7.91049 | 0.171641 | standard |
| 8.92736 | 0.171787 | standard |
| 8.93291 | 0.171782 | standard |
| 9.07475 | 0.333085 | standard |
| 4.00484 | 1.0 | standard |
| 7.90564 | 0.171901 | standard |
| 7.9099 | 0.171244 | standard |
| 8.92806 | 0.17191 | standard |
| 8.93231 | 0.171254 | standard |
| 9.07475 | 0.332664 | standard |