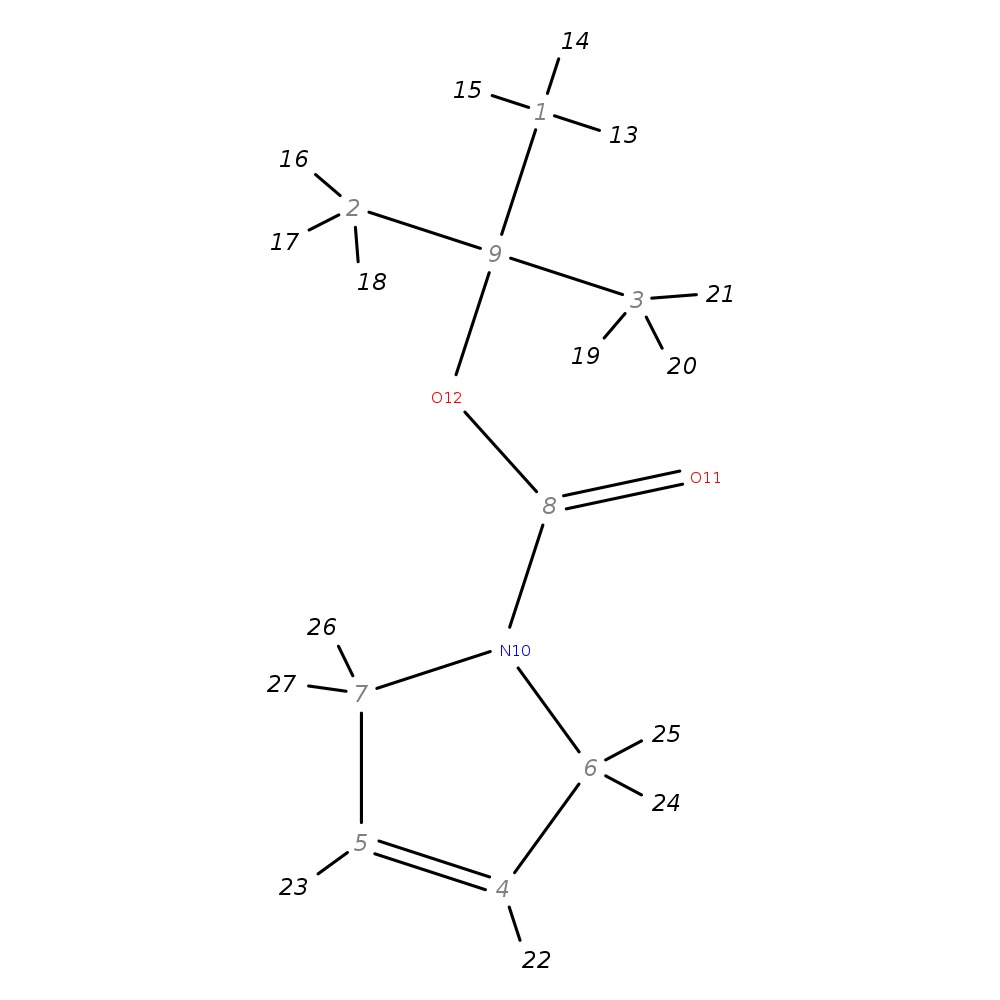
N-BOC-3-Pyrroline
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03545 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H15NO2/c1-9(2,3)12-8(11)10-6-4-5-7-10/h4-5H,6-7H2,1-3H3 | |
| Note 1 | 24,25?26,27 different shifts (QM confirmed) | |
| Note 2 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-BOC-3-Pyrroline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 1.45 | 8.9 | 8.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 1.45 | 8.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 1.452 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 1.45 | 8.9 | 8.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 1.45 | 8.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 1.452 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 1.45 | 8.9 | 8.9 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.452 | 8.9 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.45 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.982 | 7.0 | 3.98 | 3.98 | -1.0 | -1.0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.982 | -1.0 | -1.0 | 3.98 | 3.98 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.96 | 13.66 | 1.0 | 1.0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.952 | 1.0 | 1.0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.896 | 13.66 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.902 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.45962 | 1.0 | standard |
| 4.0779 | 0.0460482 | standard |
| 4.12202 | 0.0460482 | standard |
| 5.82957 | 0.044522 | standard |
| 1.45962 | 1.0 | standard |
| 4.10021 | 0.0886521 | standard |
| 5.82992 | 0.0460011 | standard |
| 1.45962 | 1.0 | standard |
| 4.10001 | 0.0845661 | standard |
| 5.82972 | 0.0453851 | standard |
| 1.45962 | 1.0 | standard |
| 4.10011 | 0.0800661 | standard |
| 5.82957 | 0.045114 | standard |
| 1.45962 | 1.0 | standard |
| 4.09996 | 0.077648 | standard |
| 5.82972 | 0.0450271 | standard |
| 1.45962 | 1.0 | standard |
| 4.10021 | 0.0751381 | standard |
| 5.82957 | 0.044964 | standard |
| 1.45962 | 1.0 | standard |
| 4.08091 | 0.056061 | standard |
| 4.11895 | 0.0560611 | standard |
| 5.82957 | 0.044673 | standard |
| 1.45962 | 1.0 | standard |
| 4.07855 | 0.050221 | standard |
| 4.12137 | 0.050221 | standard |
| 5.82957 | 0.044602 | standard |
| 1.45962 | 1.0 | standard |
| 4.07809 | 0.0478718 | standard |
| 4.12183 | 0.0478718 | standard |
| 5.82957 | 0.044561 | standard |
| 1.45962 | 1.0 | standard |
| 4.07794 | 0.046706 | standard |
| 4.12197 | 0.0467061 | standard |
| 5.82957 | 0.044537 | standard |
| 1.45962 | 1.0 | standard |
| 4.0779 | 0.0460482 | standard |
| 4.12203 | 0.0460486 | standard |
| 5.82957 | 0.044522 | standard |
| 1.45962 | 1.0 | standard |
| 4.07787 | 0.0456427 | standard |
| 4.12205 | 0.0456427 | standard |
| 5.82957 | 0.044509 | standard |
| 1.45962 | 1.0 | standard |
| 4.07786 | 0.0454945 | standard |
| 4.12206 | 0.0454945 | standard |
| 5.82957 | 0.044506 | standard |
| 1.45962 | 1.0 | standard |
| 4.07786 | 0.0453724 | standard |
| 4.12206 | 0.0453724 | standard |
| 5.82957 | 0.044502 | standard |
| 1.45962 | 1.0 | standard |
| 4.07785 | 0.0451862 | standard |
| 4.12207 | 0.0451862 | standard |
| 5.82957 | 0.044495 | standard |
| 1.45962 | 1.0 | standard |
| 4.07785 | 0.0451131 | standard |
| 4.12207 | 0.0451131 | standard |
| 5.82957 | 0.044494 | standard |
| 1.45962 | 1.0 | standard |
| 4.07785 | 0.0450511 | standard |
| 4.12207 | 0.0450511 | standard |
| 5.82957 | 0.04449 | standard |
| 1.45962 | 1.0 | standard |
| 4.07784 | 0.044812 | standard |
| 4.12207 | 0.044812 | standard |
| 5.82957 | 0.044481 | standard |