6-Methylquinoxaline
Simulation outputs:
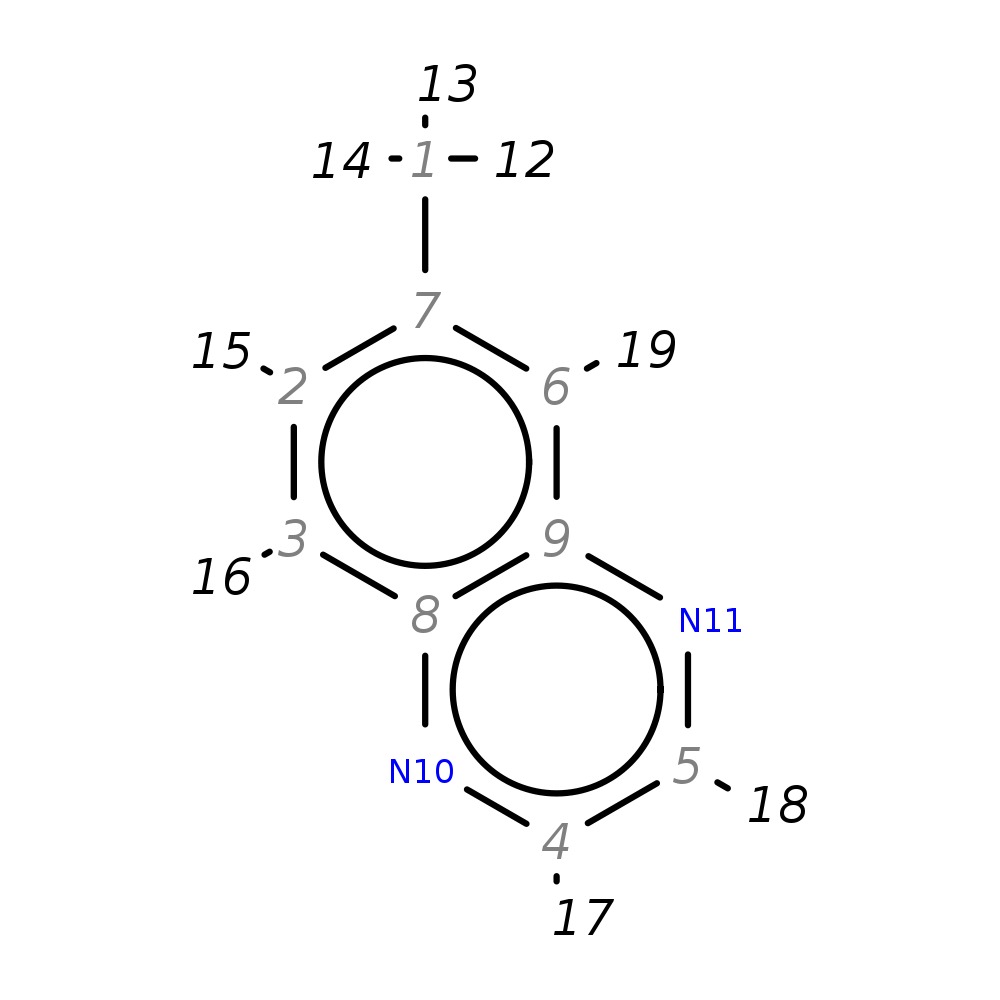
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03001 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H8N2/c1-7-2-3-8-9(6-7)11-5-4-10-8/h2-6H,1H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 6-Methylquinoxaline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|
| 12 | 2.611 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 2.611 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 2.611 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.8 | 8.831 | 0 | 0 | 1.389 |
| 16 | 0 | 0 | 0 | 0 | 8.024 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 8.858 | 1.731 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 8.835 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.928 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.61117 | 1.0 | standard |
| 7.792 | 0.110054 | standard |
| 7.79345 | 0.110482 | standard |
| 7.80661 | 0.123091 | standard |
| 7.80826 | 0.122671 | standard |
| 7.92733 | 0.226877 | standard |
| 7.92898 | 0.226879 | standard |
| 8.01732 | 0.179689 | standard |
| 8.032 | 0.15813 | standard |
| 8.83374 | 0.198254 | standard |
| 8.83614 | 0.227577 | standard |
| 8.85669 | 0.225558 | standard |
| 8.85923 | 0.195345 | standard |
| 2.61117 | 1.0 | standard |
| 7.65485 | 0.0454121 | standard |
| 7.88476 | 0.341783 | standard |
| 7.92443 | 0.4238 | standard |
| 7.95097 | 0.445738 | standard |
| 8.17855 | 0.0572411 | standard |
| 8.84642 | 0.629023 | standard |
| 2.61117 | 1.0 | standard |
| 7.71198 | 0.0618544 | standard |
| 7.85957 | 0.224564 | standard |
| 7.92192 | 0.299317 | standard |
| 7.97222 | 0.303371 | standard |
| 8.11948 | 0.080917 | standard |
| 8.84647 | 0.576508 | standard |
| 2.61117 | 1.0 | standard |
| 7.73792 | 0.072851 | standard |
| 7.84835 | 0.19055 | standard |
| 7.92279 | 0.264685 | standard |
| 7.98216 | 0.259107 | standard |
| 8.09223 | 0.0974924 | standard |
| 8.84642 | 0.504467 | standard |
| 2.61117 | 1.0 | standard |
| 7.7461 | 0.0769356 | standard |
| 7.84411 | 0.181125 | standard |
| 7.92331 | 0.256914 | standard |
| 7.98575 | 0.246969 | standard |
| 8.08357 | 0.104041 | standard |
| 8.84642 | 0.464312 | standard |
| 2.61117 | 1.0 | standard |
| 7.75243 | 0.0804026 | standard |
| 7.84062 | 0.173717 | standard |
| 7.92362 | 0.251769 | standard |
| 7.98874 | 0.238445 | standard |
| 8.07684 | 0.109473 | standard |
| 8.84518 | 0.422169 | standard |
| 8.84759 | 0.42215 | standard |
| 2.61117 | 1.0 | standard |
| 7.77402 | 0.094005 | standard |
| 7.77849 | 0.0973871 | standard |
| 7.81822 | 0.136797 | standard |
| 7.82263 | 0.142608 | standard |
| 7.92587 | 0.237571 | standard |
| 7.93006 | 0.22909 | standard |
| 8.00453 | 0.202495 | standard |
| 8.0486 | 0.138213 | standard |
| 8.83836 | 0.277173 | standard |
| 8.85447 | 0.276734 | standard |
| 2.61117 | 1.0 | standard |
| 7.78329 | 0.102207 | standard |
| 7.78618 | 0.104228 | standard |
| 7.81259 | 0.129801 | standard |
| 7.81568 | 0.133001 | standard |
| 7.92659 | 0.233219 | standard |
| 7.92957 | 0.228177 | standard |
| 8.01068 | 0.190805 | standard |
| 8.04013 | 0.147969 | standard |
| 8.83288 | 0.190165 | standard |
| 8.83728 | 0.248882 | standard |
| 8.85566 | 0.248881 | standard |
| 8.86006 | 0.190164 | standard |
| 2.61117 | 1.0 | standard |
| 7.78765 | 0.10543 | standard |
| 7.78993 | 0.106701 | standard |
| 7.80966 | 0.127404 | standard |
| 7.81194 | 0.128828 | standard |
| 7.92699 | 0.231207 | standard |
| 7.92925 | 0.228633 | standard |
| 8.01395 | 0.185226 | standard |
| 8.03602 | 0.15296 | standard |
| 8.8331 | 0.192439 | standard |
| 8.8367 | 0.236996 | standard |
| 8.85614 | 0.237 | standard |
| 8.85974 | 0.192436 | standard |
| 2.61117 | 1.0 | standard |
| 7.79029 | 0.107746 | standard |
| 7.79211 | 0.108605 | standard |
| 7.80792 | 0.125767 | standard |
| 7.80974 | 0.125914 | standard |
| 7.92723 | 0.230179 | standard |
| 7.92905 | 0.229253 | standard |
| 8.01594 | 0.181899 | standard |
| 8.03359 | 0.156055 | standard |
| 8.83343 | 0.195697 | standard |
| 8.83632 | 0.231097 | standard |
| 8.85652 | 0.231097 | standard |
| 8.85941 | 0.195696 | standard |
| 2.61117 | 1.0 | standard |
| 7.792 | 0.110055 | standard |
| 7.79345 | 0.110483 | standard |
| 7.80661 | 0.123085 | standard |
| 7.80826 | 0.122669 | standard |
| 7.92733 | 0.226876 | standard |
| 7.92898 | 0.226874 | standard |
| 8.01732 | 0.17969 | standard |
| 8.032 | 0.158123 | standard |
| 8.83374 | 0.198253 | standard |
| 8.83614 | 0.227572 | standard |
| 8.85669 | 0.225552 | standard |
| 8.85923 | 0.195344 | standard |
| 2.61117 | 1.0 | standard |
| 7.79322 | 0.110273 | standard |
| 7.79452 | 0.110812 | standard |
| 7.80579 | 0.123485 | standard |
| 7.8071 | 0.123065 | standard |
| 7.92751 | 0.22945 | standard |
| 7.9288 | 0.229524 | standard |
| 8.01832 | 0.17797 | standard |
| 8.03091 | 0.159624 | standard |
| 8.83388 | 0.19918 | standard |
| 8.83596 | 0.224551 | standard |
| 8.85688 | 0.224551 | standard |
| 8.85897 | 0.19918 | standard |
| 2.61117 | 1.0 | standard |
| 7.79367 | 0.109993 | standard |
| 7.79495 | 0.11047 | standard |
| 7.80551 | 0.124973 | standard |
| 7.80658 | 0.124594 | standard |
| 7.92762 | 0.233209 | standard |
| 7.92869 | 0.233209 | standard |
| 8.01871 | 0.177477 | standard |
| 8.03051 | 0.160211 | standard |
| 8.83396 | 0.201198 | standard |
| 8.83587 | 0.224463 | standard |
| 8.85698 | 0.224464 | standard |
| 8.85888 | 0.201198 | standard |
| 2.61117 | 1.0 | standard |
| 7.79404 | 0.110004 | standard |
| 7.79527 | 0.1104 | standard |
| 7.80519 | 0.123977 | standard |
| 7.80622 | 0.123715 | standard |
| 7.92754 | 0.22677 | standard |
| 7.92877 | 0.226734 | standard |
| 8.01901 | 0.176765 | standard |
| 8.03012 | 0.160823 | standard |
| 8.83403 | 0.200054 | standard |
| 8.83588 | 0.222213 | standard |
| 8.85706 | 0.222214 | standard |
| 8.85891 | 0.200054 | standard |
| 2.61117 | 1.0 | standard |
| 7.79486 | 0.113963 | standard |
| 7.79574 | 0.114358 | standard |
| 7.80467 | 0.123514 | standard |
| 7.80555 | 0.123154 | standard |
| 7.92761 | 0.226817 | standard |
| 7.9287 | 0.22682 | standard |
| 8.01961 | 0.175413 | standard |
| 8.02942 | 0.16222 | standard |
| 8.83409 | 0.19928 | standard |
| 8.8358 | 0.219395 | standard |
| 8.85716 | 0.222706 | standard |
| 8.85872 | 0.203471 | standard |
| 2.61117 | 1.0 | standard |
| 7.79509 | 0.111583 | standard |
| 7.7961 | 0.112057 | standard |
| 7.80441 | 0.120508 | standard |
| 7.80541 | 0.120079 | standard |
| 7.9276 | 0.227812 | standard |
| 7.92861 | 0.227802 | standard |
| 8.0199 | 0.175057 | standard |
| 8.02922 | 0.162536 | standard |
| 8.83421 | 0.203843 | standard |
| 8.83569 | 0.222095 | standard |
| 8.85714 | 0.218592 | standard |
| 8.85876 | 0.199463 | standard |
| 2.61117 | 1.0 | standard |
| 7.79539 | 0.112647 | standard |
| 7.7963 | 0.1131 | standard |
| 7.80421 | 0.121094 | standard |
| 7.80512 | 0.120737 | standard |
| 7.9277 | 0.229494 | standard |
| 7.92861 | 0.229433 | standard |
| 8.0201 | 0.174753 | standard |
| 8.02893 | 0.162761 | standard |
| 8.83417 | 0.200041 | standard |
| 8.83571 | 0.218186 | standard |
| 8.85725 | 0.221929 | standard |
| 8.85864 | 0.204609 | standard |
| 2.61117 | 1.0 | standard |
| 7.79652 | 0.113369 | standard |
| 7.79725 | 0.11384 | standard |
| 7.80341 | 0.123308 | standard |
| 7.80393 | 0.122989 | standard |
| 7.92774 | 0.227393 | standard |
| 7.92847 | 0.227393 | standard |
| 8.02109 | 0.172174 | standard |
| 8.02784 | 0.165526 | standard |
| 8.83433 | 0.201136 | standard |
| 8.83553 | 0.215109 | standard |
| 8.85743 | 0.220086 | standard |
| 8.85849 | 0.20699 | standard |