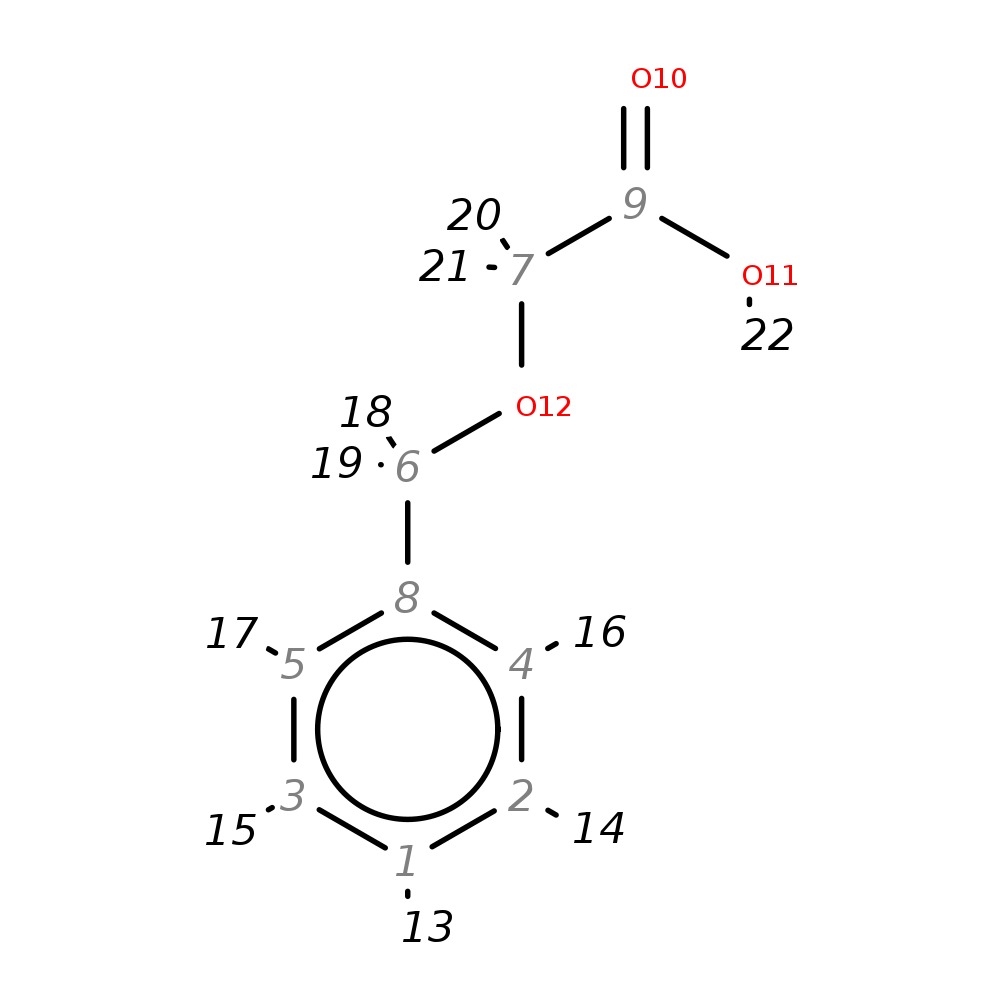
Benzyloxyacetic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.13466 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H10O3/c10-9(11)7-12-6-8-4-2-1-3-5-8/h1-5H,6-7H2,(H,10,11) | |
| Note 1 | 18,19?20,21 | |
| Note 2 | 18,19 or 2021 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Benzyloxyacetic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|
| 13 | 7.398 | 8.6 | 8.6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 7.44 | 1.5 | 7.915 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 7.44 | 0 | 7.915 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 7.44 | 1.5 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 7.44 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 4.476 | 10.59 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 4.336 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.931 | -14.0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.931 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.93124 | 0.975932 | standard |
| 4.327 | 0.216841 | standard |
| 4.34465 | 0.27556 | standard |
| 4.46814 | 0.275554 | standard |
| 4.48579 | 0.216833 | standard |
| 7.37458 | 0.00866009 | standard |
| 7.38109 | 0.0290184 | standard |
| 7.38741 | 0.0536358 | standard |
| 7.38936 | 0.0564293 | standard |
| 7.39547 | 0.113466 | standard |
| 7.40262 | 0.10383 | standard |
| 7.40986 | 0.109751 | standard |
| 7.41705 | 0.0567071 | standard |
| 7.4242 | 0.0661186 | standard |
| 7.43721 | 1.00019 | standard |
| 7.44367 | 0.567327 | standard |
| 7.45885 | 0.0289887 | standard |
| 3.93125 | 0.428363 | standard |
| 4.12302 | 0.0226571 | standard |
| 4.39502 | 0.278352 | standard |
| 4.41785 | 0.27824 | standard |
| 4.68808 | 0.0160801 | standard |
| 7.43139 | 1.0 | standard |
| 3.93124 | 0.452133 | standard |
| 4.20559 | 0.029025 | standard |
| 4.38259 | 0.223534 | standard |
| 4.43029 | 0.223423 | standard |
| 4.6071 | 0.027651 | standard |
| 7.43133 | 1.0 | standard |
| 3.93124 | 0.482238 | standard |
| 4.24395 | 0.0411882 | standard |
| 4.37639 | 0.212484 | standard |
| 4.43649 | 0.212401 | standard |
| 4.56884 | 0.0406121 | standard |
| 7.43151 | 1.0 | standard |
| 3.93124 | 0.499021 | standard |
| 4.25623 | 0.0476859 | standard |
| 4.37386 | 0.211554 | standard |
| 4.43893 | 0.211479 | standard |
| 4.55665 | 0.0472606 | standard |
| 7.4317 | 1.0 | standard |
| 3.93124 | 0.51645 | standard |
| 4.26574 | 0.0542408 | standard |
| 4.37166 | 0.211955 | standard |
| 4.44123 | 0.211889 | standard |
| 4.54705 | 0.0539229 | standard |
| 7.43192 | 1.0 | standard |
| 3.93124 | 0.705817 | standard |
| 4.30508 | 0.116637 | standard |
| 4.35804 | 0.240669 | standard |
| 4.45475 | 0.240637 | standard |
| 4.5077 | 0.116564 | standard |
| 7.34196 | 0.011932 | standard |
| 7.37724 | 0.0344502 | standard |
| 7.43401 | 1.0 | standard |
| 3.93124 | 0.855989 | standard |
| 4.31659 | 0.16462 | standard |
| 4.35189 | 0.267766 | standard |
| 4.461 | 0.267747 | standard |
| 4.4963 | 0.164586 | standard |
| 7.36762 | 0.0212718 | standard |
| 7.38832 | 0.0606855 | standard |
| 7.40278 | 0.0928233 | standard |
| 7.41131 | 0.166147 | standard |
| 7.41767 | 0.212752 | standard |
| 7.43539 | 1.0 | standard |
| 7.4805 | 0.0174732 | standard |
| 3.93124 | 0.909023 | standard |
| 4.32194 | 0.188182 | standard |
| 4.34842 | 0.270682 | standard |
| 4.46447 | 0.270682 | standard |
| 4.49085 | 0.188181 | standard |
| 7.36225 | 0.00730438 | standard |
| 7.37152 | 0.021004 | standard |
| 7.37838 | 0.0361965 | standard |
| 7.38591 | 0.046748 | standard |
| 7.39246 | 0.0831813 | standard |
| 7.40316 | 0.092107 | standard |
| 7.40762 | 0.100972 | standard |
| 7.41432 | 0.168537 | standard |
| 7.43622 | 1.00009 | standard |
| 7.44464 | 0.465329 | standard |
| 3.93124 | 0.951924 | standard |
| 4.32501 | 0.205732 | standard |
| 4.34624 | 0.274622 | standard |
| 4.46665 | 0.274629 | standard |
| 4.48778 | 0.205742 | standard |
| 7.36966 | 0.00817535 | standard |
| 7.37737 | 0.02578 | standard |
| 7.3838 | 0.0461084 | standard |
| 7.38816 | 0.0513686 | standard |
| 7.39439 | 0.101729 | standard |
| 7.40301 | 0.0987444 | standard |
| 7.40548 | 0.0949752 | standard |
| 7.41171 | 0.123853 | standard |
| 7.4203 | 0.105556 | standard |
| 7.43679 | 1.0 | standard |
| 7.4442 | 0.527787 | standard |
| 7.46281 | 0.0256712 | standard |
| 3.93124 | 0.975925 | standard |
| 4.327 | 0.21684 | standard |
| 4.34465 | 0.275559 | standard |
| 4.46814 | 0.275552 | standard |
| 4.48579 | 0.216831 | standard |
| 7.37458 | 0.00866009 | standard |
| 7.38109 | 0.0290184 | standard |
| 7.38741 | 0.0536358 | standard |
| 7.38937 | 0.0564293 | standard |
| 7.39547 | 0.113466 | standard |
| 7.40262 | 0.103831 | standard |
| 7.40986 | 0.109752 | standard |
| 7.41705 | 0.0567081 | standard |
| 7.4242 | 0.0661196 | standard |
| 7.43721 | 1.00019 | standard |
| 7.44367 | 0.567327 | standard |
| 7.45885 | 0.0289887 | standard |
| 3.93124 | 0.990144 | standard |
| 4.32849 | 0.224198 | standard |
| 4.34356 | 0.275382 | standard |
| 4.46923 | 0.275351 | standard |
| 4.4844 | 0.224161 | standard |
| 7.37764 | 0.0102561 | standard |
| 7.38375 | 0.0317182 | standard |
| 7.39028 | 0.0632595 | standard |
| 7.39608 | 0.120923 | standard |
| 7.40221 | 0.1076 | standard |
| 7.40849 | 0.0994768 | standard |
| 7.41462 | 0.046096 | standard |
| 7.4261 | 0.0574124 | standard |
| 7.43759 | 1.00001 | standard |
| 7.44322 | 0.601115 | standard |
| 7.45617 | 0.0330507 | standard |
| 3.93124 | 0.999416 | standard |
| 4.32898 | 0.227927 | standard |
| 4.34316 | 0.276259 | standard |
| 4.46973 | 0.276282 | standard |
| 4.48381 | 0.227955 | standard |
| 7.379 | 0.0106877 | standard |
| 7.38473 | 0.0324959 | standard |
| 7.39057 | 0.0680864 | standard |
| 7.39629 | 0.124755 | standard |
| 7.40207 | 0.11012 | standard |
| 7.40789 | 0.0962714 | standard |
| 7.41368 | 0.0436838 | standard |
| 7.42064 | 0.0204592 | standard |
| 7.42701 | 0.0556347 | standard |
| 7.43773 | 1.00055 | standard |
| 7.44299 | 0.613942 | standard |
| 7.45507 | 0.034276 | standard |
| 3.93124 | 1.0 | standard |
| 4.32948 | 0.229524 | standard |
| 4.34277 | 0.274956 | standard |
| 4.47012 | 0.274982 | standard |
| 4.48331 | 0.229555 | standard |
| 7.38018 | 0.0109615 | standard |
| 7.38562 | 0.0334747 | standard |
| 7.39111 | 0.0701237 | standard |
| 7.39648 | 0.126629 | standard |
| 7.40191 | 0.111249 | standard |
| 7.40738 | 0.0937628 | standard |
| 7.41278 | 0.040714 | standard |
| 7.41929 | 0.0165271 | standard |
| 7.42776 | 0.0542411 | standard |
| 7.43786 | 0.993736 | standard |
| 7.44281 | 0.625929 | standard |
| 7.45414 | 0.0348712 | standard |
| 3.93124 | 1.0 | standard |
| 4.33027 | 0.232347 | standard |
| 4.34207 | 0.272127 | standard |
| 4.47082 | 0.272158 | standard |
| 4.48252 | 0.232383 | standard |
| 7.38226 | 0.0113442 | standard |
| 7.38712 | 0.0347514 | standard |
| 7.39194 | 0.0749381 | standard |
| 7.39677 | 0.129739 | standard |
| 7.40152 | 0.112129 | standard |
| 7.40638 | 0.0861745 | standard |
| 7.41134 | 0.0371079 | standard |
| 7.41702 | 0.0123358 | standard |
| 7.42918 | 0.0508091 | standard |
| 7.43808 | 0.985938 | standard |
| 7.44252 | 0.633499 | standard |
| 7.45253 | 0.0379201 | standard |
| 3.93124 | 1.0 | standard |
| 4.33067 | 0.233635 | standard |
| 4.34177 | 0.270883 | standard |
| 4.47111 | 0.27088 | standard |
| 4.48222 | 0.233632 | standard |
| 7.38306 | 0.0117776 | standard |
| 7.38771 | 0.0354726 | standard |
| 7.39225 | 0.077421 | standard |
| 7.39687 | 0.130583 | standard |
| 7.40142 | 0.113339 | standard |
| 7.40598 | 0.0851019 | standard |
| 7.41065 | 0.0354844 | standard |
| 7.416 | 0.0110871 | standard |
| 7.42974 | 0.049969 | standard |
| 7.43817 | 0.983038 | standard |
| 7.44237 | 0.640352 | standard |
| 7.45185 | 0.0385126 | standard |
| 3.93124 | 1.0 | standard |
| 4.33096 | 0.234896 | standard |
| 4.34148 | 0.269651 | standard |
| 4.47131 | 0.26961 | standard |
| 4.48192 | 0.234849 | standard |
| 7.38385 | 0.0119532 | standard |
| 7.38831 | 0.0361201 | standard |
| 7.39261 | 0.0801659 | standard |
| 7.39697 | 0.130555 | standard |
| 7.40127 | 0.113087 | standard |
| 7.40568 | 0.0827132 | standard |
| 7.41012 | 0.0341991 | standard |
| 7.4152 | 0.0101286 | standard |
| 7.43029 | 0.0494836 | standard |
| 7.43824 | 0.989359 | standard |
| 7.44223 | 0.643883 | standard |
| 7.45127 | 0.038964 | standard |
| 3.93124 | 1.0 | standard |
| 4.33225 | 0.239774 | standard |
| 4.34039 | 0.264687 | standard |
| 4.4725 | 0.264686 | standard |
| 4.48064 | 0.239772 | standard |
| 7.38714 | 0.0131156 | standard |
| 7.3906 | 0.0394174 | standard |
| 7.39393 | 0.0883686 | standard |
| 7.39733 | 0.134796 | standard |
| 7.40062 | 0.115593 | standard |
| 7.40397 | 0.0745132 | standard |
| 7.40742 | 0.0291863 | standard |
| 7.41131 | 0.00716692 | standard |
| 7.43252 | 0.04648 | standard |
| 7.43869 | 0.984634 | standard |
| 7.44169 | 0.647136 | standard |
| 7.44867 | 0.0418903 | standard |