7-Methyl-1,2,3,4-tetrahydro-1,8-naphthyridine
Simulation outputs:
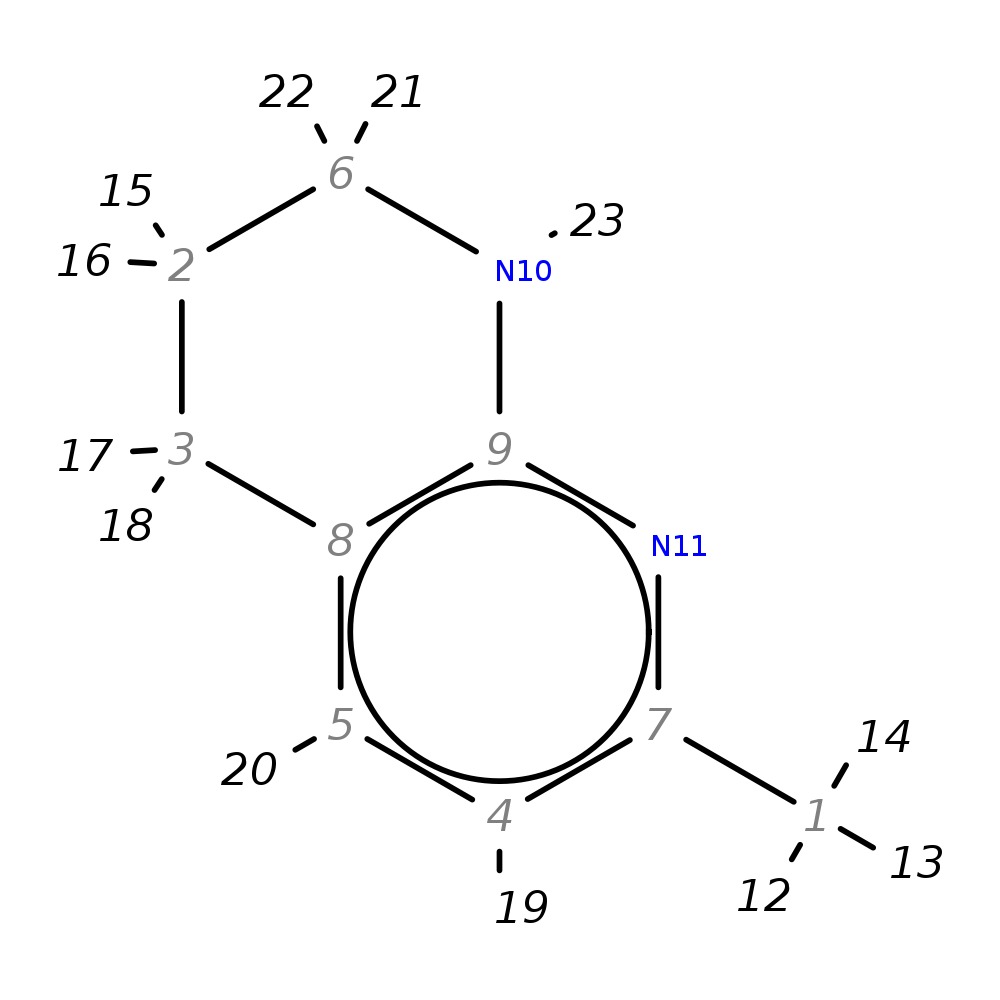
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03560 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H12N2/c1-7-4-5-8-3-2-6-10-9(8)11-7/h4-5H,2-3,6H2,1H3,(H,10,11) | |
| Note 1 | 21,22?17,18 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 7-Methyl-1,2,3,4-tetrahydro-1,8-naphthyridine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 2.356 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 2.356 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 2.356 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 1.895 | -14.0 | 6.618 | 6.618 | 0 | 0 | 5.784 | 5.784 |
| 16 | 0 | 0 | 0 | 0 | 1.895 | 6.618 | 6.618 | 0 | 0 | 5.784 | 5.784 |
| 17 | 0 | 0 | 0 | 0 | 0 | 2.749 | -14.0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 2.749 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.469 | 8.205 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.534 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.423 | -14.0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.423 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.87386 | 0.0479154 | standard |
| 1.88419 | 0.150342 | standard |
| 1.89453 | 0.224258 | standard |
| 1.90481 | 0.150401 | standard |
| 1.91515 | 0.0479224 | standard |
| 2.35577 | 1.0 | standard |
| 2.7377 | 0.174652 | standard |
| 2.74872 | 0.339455 | standard |
| 2.75974 | 0.17421 | standard |
| 3.41382 | 0.177225 | standard |
| 3.42346 | 0.342092 | standard |
| 3.43306 | 0.177215 | standard |
| 6.52695 | 0.169077 | standard |
| 6.54053 | 0.167976 | standard |
| 7.46208 | 0.169036 | standard |
| 7.47576 | 0.167936 | standard |
| 1.55251 | 0.0209131 | standard |
| 1.58802 | 0.0239881 | standard |
| 1.73203 | 0.0959321 | standard |
| 1.88366 | 0.182828 | standard |
| 2.02949 | 0.174488 | standard |
| 2.16211 | 0.0746291 | standard |
| 2.18913 | 0.08105 | standard |
| 2.35577 | 1.0 | standard |
| 2.60115 | 0.214414 | standard |
| 2.75467 | 0.255026 | standard |
| 2.91305 | 0.0927981 | standard |
| 3.29025 | 0.197897 | standard |
| 3.42856 | 0.31683 | standard |
| 3.57154 | 0.137649 | standard |
| 6.4202 | 0.132774 | standard |
| 6.62521 | 0.202517 | standard |
| 7.37749 | 0.202469 | standard |
| 7.58251 | 0.131845 | standard |
| 1.68611 | 0.0285981 | standard |
| 1.785 | 0.113618 | standard |
| 1.88758 | 0.19611 | standard |
| 1.98877 | 0.167956 | standard |
| 2.08722 | 0.0583671 | standard |
| 2.35577 | 1.0 | standard |
| 2.64645 | 0.199453 | standard |
| 2.75403 | 0.286423 | standard |
| 2.8632 | 0.1157 | standard |
| 3.33142 | 0.19299 | standard |
| 3.4262 | 0.331893 | standard |
| 3.52177 | 0.152098 | standard |
| 6.46043 | 0.145059 | standard |
| 6.59715 | 0.192489 | standard |
| 7.40556 | 0.192488 | standard |
| 7.54218 | 0.145042 | standard |
| 1.7362 | 0.0327751 | standard |
| 1.81288 | 0.124267 | standard |
| 1.88982 | 0.205451 | standard |
| 1.9668 | 0.165006 | standard |
| 2.04235 | 0.055725 | standard |
| 2.35577 | 1.0 | standard |
| 2.67044 | 0.195759 | standard |
| 2.7523 | 0.306246 | standard |
| 2.83451 | 0.131908 | standard |
| 3.35346 | 0.19078 | standard |
| 3.42506 | 0.33613 | standard |
| 3.49689 | 0.159444 | standard |
| 6.47976 | 0.150943 | standard |
| 6.58228 | 0.186829 | standard |
| 7.42044 | 0.18682 | standard |
| 7.52295 | 0.150966 | standard |
| 1.75376 | 0.0344381 | standard |
| 1.82228 | 0.127639 | standard |
| 1.89066 | 0.208793 | standard |
| 1.95925 | 0.164053 | standard |
| 2.02675 | 0.054964 | standard |
| 2.35577 | 1.0 | standard |
| 2.67867 | 0.194576 | standard |
| 2.75163 | 0.312755 | standard |
| 2.82478 | 0.137619 | standard |
| 3.36094 | 0.189852 | standard |
| 3.42472 | 0.337495 | standard |
| 3.48864 | 0.162023 | standard |
| 6.48601 | 0.153247 | standard |
| 6.57712 | 0.185119 | standard |
| 7.42559 | 0.185125 | standard |
| 7.5167 | 0.153232 | standard |
| 1.76797 | 0.035735 | standard |
| 1.82979 | 0.130317 | standard |
| 1.89133 | 0.211486 | standard |
| 1.95317 | 0.162919 | standard |
| 2.01414 | 0.0544252 | standard |
| 2.35577 | 1.0 | standard |
| 2.68534 | 0.192926 | standard |
| 2.7511 | 0.317504 | standard |
| 2.81698 | 0.141693 | standard |
| 3.36702 | 0.188633 | standard |
| 3.42447 | 0.338169 | standard |
| 3.48201 | 0.163502 | standard |
| 6.49096 | 0.154644 | standard |
| 6.57296 | 0.182741 | standard |
| 7.42966 | 0.183333 | standard |
| 7.51175 | 0.154148 | standard |
| 1.83192 | 0.0431192 | standard |
| 1.86301 | 0.144226 | standard |
| 1.89377 | 0.221306 | standard |
| 1.92478 | 0.155292 | standard |
| 1.95552 | 0.0513087 | standard |
| 2.35577 | 1.0 | standard |
| 2.71626 | 0.185712 | standard |
| 2.74928 | 0.334624 | standard |
| 2.7823 | 0.159951 | standard |
| 3.39486 | 0.178615 | standard |
| 3.42368 | 0.341791 | standard |
| 3.45254 | 0.175419 | standard |
| 6.51287 | 0.162472 | standard |
| 6.55382 | 0.175419 | standard |
| 7.44889 | 0.175431 | standard |
| 7.48984 | 0.162471 | standard |
| 1.85297 | 0.0455065 | standard |
| 1.87366 | 0.14855 | standard |
| 1.89425 | 0.22363 | standard |
| 1.91485 | 0.152646 | standard |
| 1.93544 | 0.0498909 | standard |
| 2.35577 | 1.0 | standard |
| 2.72691 | 0.180499 | standard |
| 2.74893 | 0.33771 | standard |
| 2.77093 | 0.167473 | standard |
| 3.4043 | 0.177371 | standard |
| 3.42353 | 0.342395 | standard |
| 3.44276 | 0.177191 | standard |
| 6.51991 | 0.165824 | standard |
| 6.54728 | 0.172152 | standard |
| 7.45544 | 0.172268 | standard |
| 7.4828 | 0.165721 | standard |
| 1.86346 | 0.0470795 | standard |
| 1.87895 | 0.150301 | standard |
| 1.8944 | 0.223375 | standard |
| 1.90983 | 0.150468 | standard |
| 1.92535 | 0.0495215 | standard |
| 2.35577 | 1.0 | standard |
| 2.73227 | 0.17655 | standard |
| 2.7488 | 0.33911 | standard |
| 2.76534 | 0.172734 | standard |
| 3.40905 | 0.177196 | standard |
| 3.42346 | 0.342541 | standard |
| 3.43791 | 0.177345 | standard |
| 6.52338 | 0.167925 | standard |
| 6.54391 | 0.17002 | standard |
| 7.45881 | 0.170204 | standard |
| 7.47923 | 0.168325 | standard |
| 1.86975 | 0.0473241 | standard |
| 1.88209 | 0.150009 | standard |
| 1.89451 | 0.224509 | standard |
| 1.90681 | 0.150127 | standard |
| 1.91925 | 0.0485384 | standard |
| 2.35577 | 1.0 | standard |
| 2.73557 | 0.175974 | standard |
| 2.74874 | 0.339955 | standard |
| 2.76197 | 0.172853 | standard |
| 3.41187 | 0.177391 | standard |
| 3.42346 | 0.342679 | standard |
| 3.43504 | 0.177392 | standard |
| 6.52546 | 0.169072 | standard |
| 6.54192 | 0.169033 | standard |
| 7.46079 | 0.169041 | standard |
| 7.47715 | 0.169111 | standard |
| 1.87386 | 0.0479814 | standard |
| 1.88419 | 0.15055 | standard |
| 1.89453 | 0.224522 | standard |
| 1.90481 | 0.150609 | standard |
| 1.91515 | 0.0479884 | standard |
| 2.35577 | 1.0 | standard |
| 2.7377 | 0.174893 | standard |
| 2.74872 | 0.339899 | standard |
| 2.75974 | 0.174451 | standard |
| 3.41382 | 0.177474 | standard |
| 3.42346 | 0.342663 | standard |
| 3.43306 | 0.177464 | standard |
| 6.52695 | 0.169108 | standard |
| 6.54053 | 0.16912 | standard |
| 7.46208 | 0.169087 | standard |
| 7.47576 | 0.169069 | standard |
| 1.87688 | 0.0480487 | standard |
| 1.8857 | 0.150423 | standard |
| 1.89455 | 0.224182 | standard |
| 1.90338 | 0.150329 | standard |
| 1.91223 | 0.0480249 | standard |
| 2.35577 | 1.0 | standard |
| 2.73928 | 0.174649 | standard |
| 2.74869 | 0.339868 | standard |
| 2.75811 | 0.174648 | standard |
| 3.41519 | 0.177293 | standard |
| 3.42345 | 0.342078 | standard |
| 3.43168 | 0.177176 | standard |
| 6.52784 | 0.16789 | standard |
| 6.53964 | 0.168998 | standard |
| 7.46307 | 0.169094 | standard |
| 7.47477 | 0.16787 | standard |
| 1.87806 | 0.0481008 | standard |
| 1.88632 | 0.149536 | standard |
| 1.89455 | 0.225225 | standard |
| 1.90278 | 0.149537 | standard |
| 1.91104 | 0.0481018 | standard |
| 2.35577 | 1.0 | standard |
| 2.73988 | 0.174822 | standard |
| 2.74869 | 0.340173 | standard |
| 2.75751 | 0.174821 | standard |
| 3.41574 | 0.177422 | standard |
| 3.42341 | 0.343 | standard |
| 3.43113 | 0.177534 | standard |
| 6.52824 | 0.169072 | standard |
| 6.53925 | 0.169075 | standard |
| 7.46347 | 0.169118 | standard |
| 7.47437 | 0.169117 | standard |
| 1.87905 | 0.0480285 | standard |
| 1.88683 | 0.149609 | standard |
| 1.89458 | 0.224716 | standard |
| 1.90229 | 0.150586 | standard |
| 1.91005 | 0.0480483 | standard |
| 2.35577 | 1.0 | standard |
| 2.74042 | 0.175005 | standard |
| 2.74869 | 0.340171 | standard |
| 2.75692 | 0.174827 | standard |
| 3.41623 | 0.177389 | standard |
| 3.42341 | 0.342947 | standard |
| 3.43064 | 0.177524 | standard |
| 6.52864 | 0.168961 | standard |
| 6.53885 | 0.169122 | standard |
| 7.46377 | 0.168911 | standard |
| 7.47408 | 0.169074 | standard |
| 1.88084 | 0.0480352 | standard |
| 1.88771 | 0.150934 | standard |
| 1.89457 | 0.224901 | standard |
| 1.90144 | 0.149835 | standard |
| 1.90836 | 0.0479798 | standard |
| 2.35577 | 1.0 | standard |
| 2.74136 | 0.174638 | standard |
| 2.74869 | 0.339879 | standard |
| 2.75603 | 0.174638 | standard |
| 3.41702 | 0.177224 | standard |
| 3.42341 | 0.342762 | standard |
| 3.42984 | 0.177414 | standard |
| 6.52923 | 0.169074 | standard |
| 6.53835 | 0.168545 | standard |
| 7.46436 | 0.169091 | standard |
| 7.47348 | 0.168601 | standard |
| 1.88153 | 0.0480191 | standard |
| 1.88806 | 0.151073 | standard |
| 1.89458 | 0.224745 | standard |
| 1.90108 | 0.149867 | standard |
| 1.90757 | 0.0480864 | standard |
| 2.35577 | 1.0 | standard |
| 2.74171 | 0.174968 | standard |
| 2.74869 | 0.339624 | standard |
| 2.75563 | 0.174696 | standard |
| 3.41737 | 0.177685 | standard |
| 3.42341 | 0.342979 | standard |
| 3.4295 | 0.177154 | standard |
| 6.52943 | 0.168931 | standard |
| 6.53806 | 0.169009 | standard |
| 7.46456 | 0.169033 | standard |
| 7.47328 | 0.168589 | standard |
| 1.88222 | 0.0480755 | standard |
| 1.8884 | 0.150181 | standard |
| 1.89457 | 0.224158 | standard |
| 1.90076 | 0.150185 | standard |
| 1.90698 | 0.0479965 | standard |
| 2.35577 | 1.0 | standard |
| 2.74206 | 0.174576 | standard |
| 2.74869 | 0.339926 | standard |
| 2.75528 | 0.175101 | standard |
| 3.41767 | 0.177689 | standard |
| 3.42341 | 0.342994 | standard |
| 3.4292 | 0.177109 | standard |
| 6.52963 | 0.169086 | standard |
| 6.53786 | 0.16909 | standard |
| 7.46486 | 0.169149 | standard |
| 7.47299 | 0.169148 | standard |
| 1.88504 | 0.0480049 | standard |
| 1.88984 | 0.148525 | standard |
| 1.8946 | 0.223658 | standard |
| 1.89936 | 0.148546 | standard |
| 1.90411 | 0.0479367 | standard |
| 2.35577 | 1.0 | standard |
| 2.74359 | 0.174894 | standard |
| 2.74864 | 0.340277 | standard |
| 2.75375 | 0.174039 | standard |
| 3.41895 | 0.177259 | standard |
| 3.42341 | 0.342426 | standard |
| 3.42786 | 0.177259 | standard |
| 6.53062 | 0.169059 | standard |
| 6.53697 | 0.167753 | standard |
| 7.46575 | 0.169035 | standard |
| 7.47209 | 0.167994 | standard |