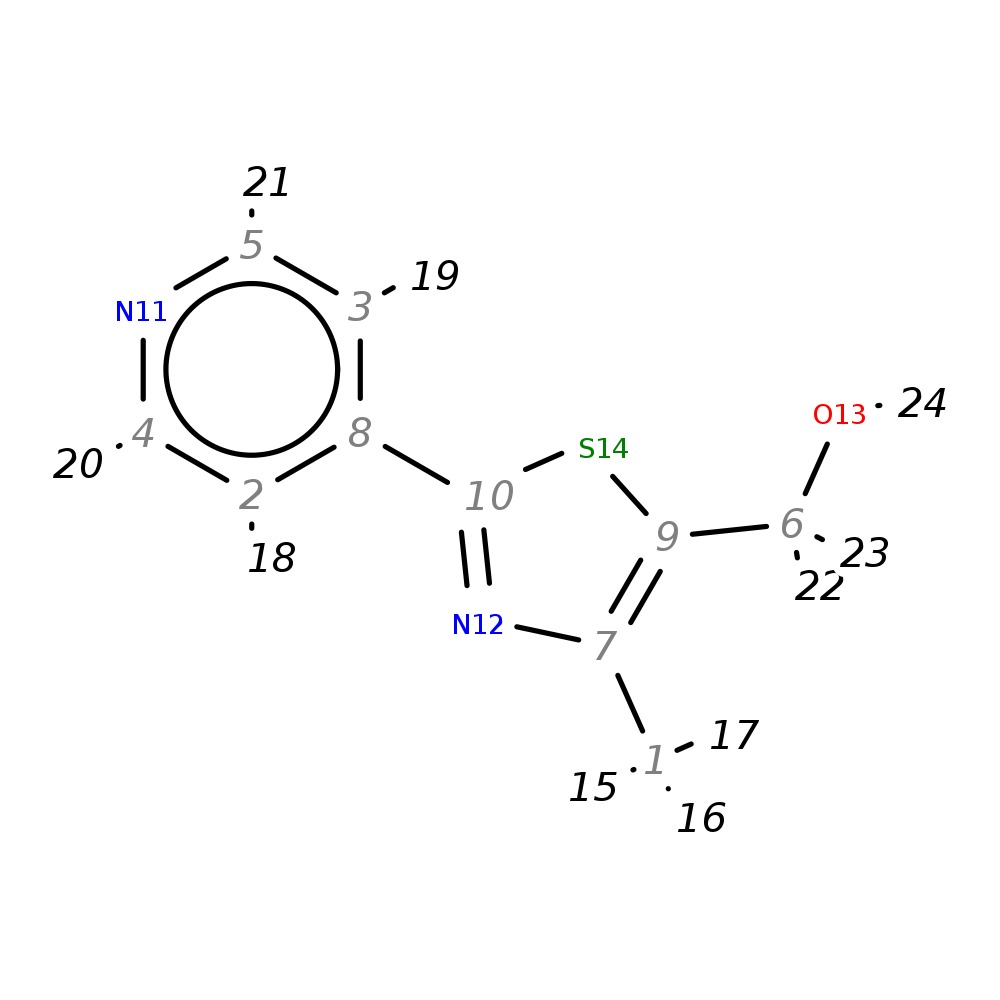
(4-Methyl-2-pyrid-4-yl-1,3-thiazol-5-yl)methanol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.20509 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H10N2OS/c1-7-9(6-13)14-10(12-7)8-2-4-11-5-3-8/h2-5,13H,6H2,1H3 | |
| Note 1 | 22,23 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| (4-Methyl-2-pyrid-4-yl-1,3-thiazol-5-yl)methanol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|
| 15 | 2.452 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 2.452 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 2.452 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.867 | 1.5 | 5.72 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 7.867 | 0 | 5.72 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 8.636 | 1.5 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 8.636 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.867 | 13.08 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.867 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666642 | standard |
| 7.86269 | 0.293208 | standard |
| 7.87202 | 0.293204 | standard |
| 8.63098 | 0.293203 | standard |
| 8.64032 | 0.292975 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666694 | standard |
| 7.79067 | 0.24395 | standard |
| 7.93132 | 0.34079 | standard |
| 8.57175 | 0.340847 | standard |
| 8.71234 | 0.24392 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666587 | standard |
| 7.81771 | 0.259762 | standard |
| 7.9113 | 0.325695 | standard |
| 8.5917 | 0.325504 | standard |
| 8.68529 | 0.259677 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666523 | standard |
| 7.83067 | 0.268483 | standard |
| 7.90084 | 0.31701 | standard |
| 8.60216 | 0.317061 | standard |
| 8.67233 | 0.268433 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666614 | standard |
| 7.83494 | 0.271041 | standard |
| 7.89724 | 0.31468 | standard |
| 8.60576 | 0.31444 | standard |
| 8.66805 | 0.271253 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666456 | standard |
| 7.83828 | 0.273303 | standard |
| 7.89438 | 0.312656 | standard |
| 8.60862 | 0.312645 | standard |
| 8.66472 | 0.27328 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666571 | standard |
| 7.85306 | 0.289187 | standard |
| 7.88116 | 0.296815 | standard |
| 8.62184 | 0.296831 | standard |
| 8.64994 | 0.289175 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666522 | standard |
| 7.85792 | 0.292527 | standard |
| 7.87659 | 0.293511 | standard |
| 8.62642 | 0.293563 | standard |
| 8.64508 | 0.292475 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666602 | standard |
| 7.86036 | 0.293517 | standard |
| 7.87436 | 0.292586 | standard |
| 8.62865 | 0.292482 | standard |
| 8.64275 | 0.292372 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666608 | standard |
| 7.86171 | 0.29278 | standard |
| 7.87299 | 0.292733 | standard |
| 8.63012 | 0.294166 | standard |
| 8.64129 | 0.292651 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666632 | standard |
| 7.86269 | 0.293178 | standard |
| 7.87202 | 0.293154 | standard |
| 8.63098 | 0.293166 | standard |
| 8.64032 | 0.292947 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666626 | standard |
| 7.86337 | 0.293628 | standard |
| 7.87134 | 0.293678 | standard |
| 8.63167 | 0.293613 | standard |
| 8.63963 | 0.293606 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666694 | standard |
| 7.86366 | 0.293557 | standard |
| 7.87115 | 0.291416 | standard |
| 8.63196 | 0.293469 | standard |
| 8.63934 | 0.293459 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666543 | standard |
| 7.86386 | 0.293676 | standard |
| 7.87085 | 0.293442 | standard |
| 8.63215 | 0.293433 | standard |
| 8.63915 | 0.293344 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666595 | standard |
| 7.86424 | 0.292137 | standard |
| 7.87046 | 0.294806 | standard |
| 8.63254 | 0.29224 | standard |
| 8.63876 | 0.292239 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666609 | standard |
| 7.86444 | 0.292817 | standard |
| 7.87037 | 0.293172 | standard |
| 8.63274 | 0.293171 | standard |
| 8.63857 | 0.293164 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666617 | standard |
| 7.86453 | 0.291466 | standard |
| 7.87017 | 0.293968 | standard |
| 8.63283 | 0.291276 | standard |
| 8.63847 | 0.291195 | standard |
| 2.45218 | 1.0 | standard |
| 4.86687 | 0.666606 | standard |
| 7.86521 | 0.292522 | standard |
| 7.86949 | 0.295888 | standard |
| 8.63351 | 0.29219 | standard |
| 8.63779 | 0.292174 | standard |