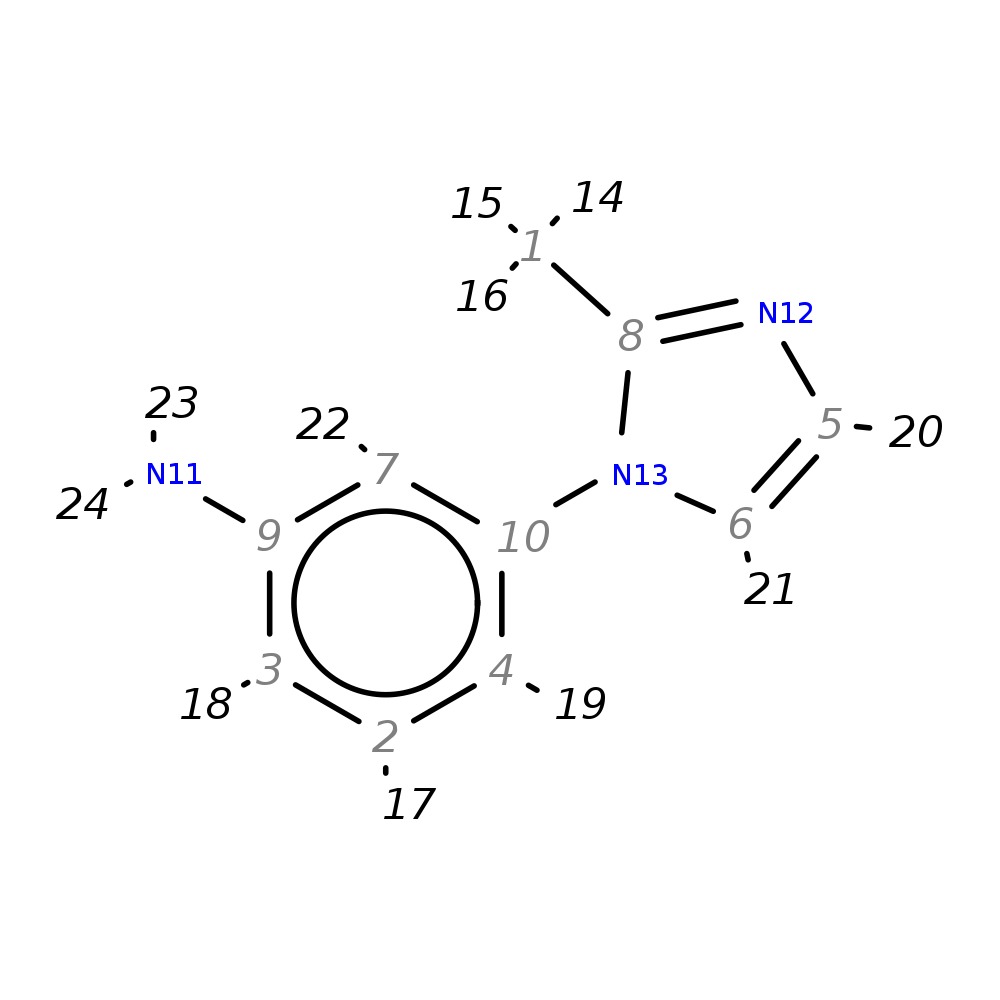
3-(2-Methyl-1H-imidazol-1-yl)aniline
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02840 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H11N3/c1-8-12-5-6-13(8)10-4-2-3-9(11)7-10/h2-7H,11H2,1H3 | |
| Note 1 | 18?19 | |
| Note 2 | 20?21 (broad peaks) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(2-Methyl-1H-imidazol-1-yl)aniline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 2.419 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.419 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.419 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.371 | 8.291 | 8.291 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 6.962 | 1.5 | 0 | 0 | 1.5 |
| 19 | 0 | 0 | 0 | 0 | 0 | 6.854 | 0 | 0 | 1.505 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.321 | 0.08 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.183 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.852 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.41905 | 1.0 | standard |
| 6.84972 | 0.250454 | standard |
| 6.85148 | 0.256843 | standard |
| 6.85902 | 0.106574 | standard |
| 6.86109 | 0.123458 | standard |
| 6.95554 | 0.109312 | standard |
| 6.96931 | 0.114278 | standard |
| 7.18331 | 0.33324 | standard |
| 7.32064 | 0.332722 | standard |
| 7.35742 | 0.0826256 | standard |
| 7.37142 | 0.153213 | standard |
| 7.38547 | 0.0759488 | standard |
| 2.41905 | 1.0 | standard |
| 6.73688 | 0.0879422 | standard |
| 6.86924 | 0.339617 | standard |
| 6.93215 | 0.218317 | standard |
| 7.00934 | 0.211684 | standard |
| 7.18642 | 0.434907 | standard |
| 7.32014 | 0.384153 | standard |
| 7.39855 | 0.116147 | standard |
| 7.6118 | 0.0336211 | standard |
| 2.41905 | 1.0 | standard |
| 6.7785 | 0.100324 | standard |
| 6.85483 | 0.299168 | standard |
| 6.87124 | 0.299253 | standard |
| 6.9091 | 0.21448 | standard |
| 7.00078 | 0.146023 | standard |
| 7.01916 | 0.151013 | standard |
| 7.18343 | 0.350808 | standard |
| 7.25639 | 0.170576 | standard |
| 7.32058 | 0.366146 | standard |
| 7.39057 | 0.152368 | standard |
| 7.52769 | 0.0468404 | standard |
| 2.41905 | 1.0 | standard |
| 6.79861 | 0.109015 | standard |
| 6.85171 | 0.254829 | standard |
| 6.86875 | 0.236622 | standard |
| 6.90115 | 0.236545 | standard |
| 7.00822 | 0.139818 | standard |
| 7.18338 | 0.340441 | standard |
| 7.28044 | 0.155815 | standard |
| 7.32068 | 0.35907 | standard |
| 7.38236 | 0.159737 | standard |
| 7.48637 | 0.053348 | standard |
| 2.41905 | 1.0 | standard |
| 6.80517 | 0.111681 | standard |
| 6.85168 | 0.250091 | standard |
| 6.89619 | 0.22465 | standard |
| 7.0038 | 0.136682 | standard |
| 7.18332 | 0.338406 | standard |
| 7.28953 | 0.157407 | standard |
| 7.32056 | 0.357154 | standard |
| 7.38008 | 0.159863 | standard |
| 7.42325 | 0.0257461 | standard |
| 7.4728 | 0.0556951 | standard |
| 2.41905 | 1.0 | standard |
| 6.8104 | 0.114099 | standard |
| 6.85168 | 0.24769 | standard |
| 6.89366 | 0.188206 | standard |
| 7.0002 | 0.134012 | standard |
| 7.18337 | 0.337349 | standard |
| 7.29749 | 0.164743 | standard |
| 7.32053 | 0.357783 | standard |
| 7.37844 | 0.159782 | standard |
| 7.41739 | 0.0256241 | standard |
| 7.46203 | 0.0577651 | standard |
| 2.41905 | 1.0 | standard |
| 6.83324 | 0.128668 | standard |
| 6.85159 | 0.242543 | standard |
| 6.87405 | 0.140942 | standard |
| 6.94091 | 0.103028 | standard |
| 6.98231 | 0.122538 | standard |
| 7.18336 | 0.33383 | standard |
| 7.32075 | 0.352837 | standard |
| 7.35173 | 0.0315734 | standard |
| 7.37306 | 0.157326 | standard |
| 7.41506 | 0.067233 | standard |
| 2.41905 | 1.0 | standard |
| 6.85158 | 0.243704 | standard |
| 6.86302 | 0.122829 | standard |
| 6.86764 | 0.134146 | standard |
| 6.94831 | 0.105267 | standard |
| 6.97595 | 0.118926 | standard |
| 7.18331 | 0.333034 | standard |
| 7.32064 | 0.335853 | standard |
| 7.34405 | 0.097507 | standard |
| 7.37205 | 0.155596 | standard |
| 7.40008 | 0.0712035 | standard |
| 2.41905 | 1.0 | standard |
| 6.84824 | 0.219112 | standard |
| 6.85161 | 0.246107 | standard |
| 6.86117 | 0.116195 | standard |
| 6.86436 | 0.129926 | standard |
| 6.95195 | 0.106898 | standard |
| 6.97266 | 0.116712 | standard |
| 7.18336 | 0.332853 | standard |
| 7.32064 | 0.333852 | standard |
| 7.35067 | 0.0895585 | standard |
| 7.35887 | 0.0214749 | standard |
| 7.3717 | 0.155176 | standard |
| 7.38395 | 0.0194254 | standard |
| 7.39275 | 0.0733153 | standard |
| 2.41905 | 1.0 | standard |
| 6.84902 | 0.235985 | standard |
| 6.85153 | 0.250206 | standard |
| 6.86 | 0.112079 | standard |
| 6.86232 | 0.126774 | standard |
| 6.95409 | 0.107251 | standard |
| 6.9707 | 0.115305 | standard |
| 7.18331 | 0.33325 | standard |
| 7.32064 | 0.333196 | standard |
| 7.35469 | 0.0853534 | standard |
| 7.37152 | 0.153733 | standard |
| 7.38244 | 0.0197381 | standard |
| 7.38834 | 0.0750148 | standard |
| 2.41905 | 1.0 | standard |
| 6.84972 | 0.250464 | standard |
| 6.85148 | 0.256857 | standard |
| 6.85902 | 0.106535 | standard |
| 6.86109 | 0.123432 | standard |
| 6.95554 | 0.109321 | standard |
| 6.96931 | 0.114226 | standard |
| 7.18331 | 0.333203 | standard |
| 7.32064 | 0.332685 | standard |
| 7.35742 | 0.0826206 | standard |
| 7.37142 | 0.153224 | standard |
| 7.38547 | 0.0759487 | standard |
| 2.41905 | 1.0 | standard |
| 6.85014 | 0.262598 | standard |
| 6.85139 | 0.26306 | standard |
| 6.86012 | 0.122238 | standard |
| 6.95653 | 0.109126 | standard |
| 6.96837 | 0.113377 | standard |
| 7.18331 | 0.333038 | standard |
| 7.32064 | 0.3321 | standard |
| 7.35935 | 0.0814655 | standard |
| 7.37141 | 0.151764 | standard |
| 7.38344 | 0.0766084 | standard |
| 2.41905 | 1.0 | standard |
| 6.85044 | 0.266675 | standard |
| 6.8513 | 0.26758 | standard |
| 6.85816 | 0.102265 | standard |
| 6.85974 | 0.120924 | standard |
| 6.95693 | 0.110742 | standard |
| 6.96803 | 0.111947 | standard |
| 7.18331 | 0.332917 | standard |
| 7.32059 | 0.333144 | standard |
| 7.36014 | 0.0803749 | standard |
| 7.37134 | 0.150818 | standard |
| 7.3826 | 0.0773088 | standard |
| 2.41905 | 1.0 | standard |
| 6.85129 | 0.275188 | standard |
| 6.85934 | 0.121141 | standard |
| 6.95732 | 0.110459 | standard |
| 6.96763 | 0.112202 | standard |
| 7.18331 | 0.332653 | standard |
| 7.32059 | 0.332844 | standard |
| 7.36084 | 0.0797714 | standard |
| 7.37132 | 0.150952 | standard |
| 7.3819 | 0.0774566 | standard |
| 2.41905 | 1.0 | standard |
| 6.85131 | 0.288011 | standard |
| 6.85875 | 0.1198 | standard |
| 6.95793 | 0.111728 | standard |
| 6.96708 | 0.112528 | standard |
| 7.18331 | 0.332594 | standard |
| 7.32059 | 0.332742 | standard |
| 7.36194 | 0.0777882 | standard |
| 7.37132 | 0.148843 | standard |
| 7.38071 | 0.07676 | standard |
| 2.41905 | 1.0 | standard |
| 6.85138 | 0.29623 | standard |
| 6.85856 | 0.119585 | standard |
| 6.95812 | 0.111052 | standard |
| 6.96684 | 0.111925 | standard |
| 7.18331 | 0.3326 | standard |
| 7.32064 | 0.333484 | standard |
| 7.36244 | 0.0772376 | standard |
| 7.37132 | 0.147595 | standard |
| 7.38021 | 0.0763777 | standard |
| 2.41905 | 1.0 | standard |
| 6.85143 | 0.298953 | standard |
| 6.85838 | 0.118143 | standard |
| 6.95831 | 0.111697 | standard |
| 6.96664 | 0.112132 | standard |
| 7.18331 | 0.33252 | standard |
| 7.32064 | 0.333473 | standard |
| 7.36288 | 0.0778543 | standard |
| 7.37132 | 0.146539 | standard |
| 7.37971 | 0.0759623 | standard |
| 2.41905 | 1.0 | standard |
| 6.85166 | 0.325862 | standard |
| 6.8574 | 0.116546 | standard |
| 6.95931 | 0.110438 | standard |
| 6.96564 | 0.111939 | standard |
| 7.18331 | 0.331838 | standard |
| 7.32064 | 0.333324 | standard |
| 7.36477 | 0.0722783 | standard |
| 7.37132 | 0.142774 | standard |
| 7.37777 | 0.0722716 | standard |