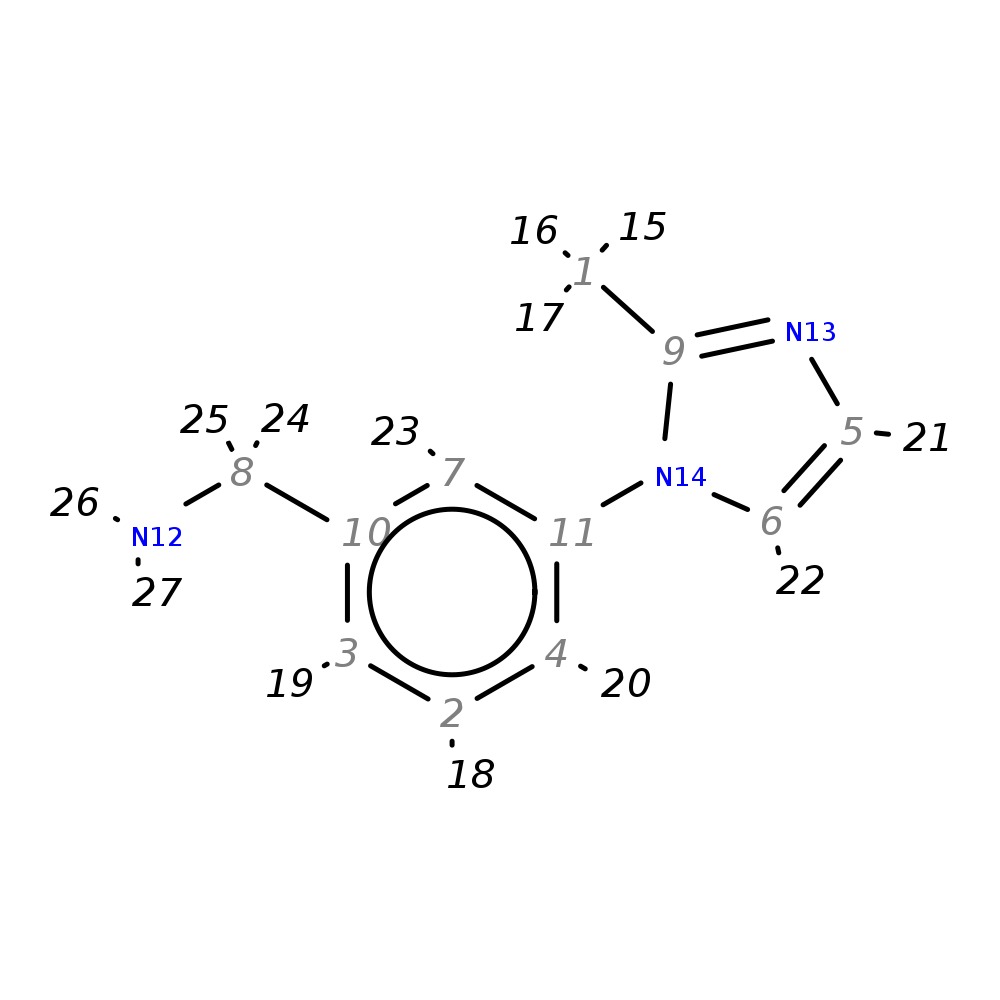
3-(2-Methyl-1H-imidazol-1-yl)benzylamine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02038 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H13N3/c1-9-13-5-6-14(9)11-4-2-3-10(7-11)8-12/h2-7H,8,12H2,1H3 | |
| Note 1 | 19?20 | |
| Note 2 | 21?22 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(2-Methyl-1H-imidazol-1-yl)benzylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 2.378 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 2.378 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 2.378 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.657 | 8.456 | 8.456 | 0 | 0 | 0.5 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 7.511 | 0 | 0 | 0 | 1.5 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 7.604 | 0 | 0 | 1.5 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 7.137 | 0.084 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.315 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.517 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.268 | -14.0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.268 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.666442 | standard |
| 7.13729 | 0.332997 | standard |
| 7.31538 | 0.332164 | standard |
| 7.50511 | 0.119268 | standard |
| 7.51754 | 0.382145 | standard |
| 7.59508 | 0.0863026 | standard |
| 7.59685 | 0.0843605 | standard |
| 7.61002 | 0.157179 | standard |
| 7.64376 | 0.106112 | standard |
| 7.65824 | 0.134884 | standard |
| 7.6723 | 0.0484397 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.666822 | standard |
| 7.13738 | 0.345588 | standard |
| 7.31545 | 0.358705 | standard |
| 7.55751 | 0.541468 | standard |
| 7.59259 | 0.615071 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.664397 | standard |
| 7.13731 | 0.338375 | standard |
| 7.3154 | 0.343491 | standard |
| 7.53655 | 0.406583 | standard |
| 7.60308 | 0.47219 | standard |
| 7.71277 | 0.0431211 | standard |
| 7.8439 | 0.0116231 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.666299 | standard |
| 7.1373 | 0.336311 | standard |
| 7.31544 | 0.338989 | standard |
| 7.43564 | 0.0464183 | standard |
| 7.53585 | 0.347547 | standard |
| 7.61018 | 0.424848 | standard |
| 7.68569 | 0.0491611 | standard |
| 7.70528 | 0.051602 | standard |
| 7.79539 | 0.0140632 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.664588 | standard |
| 7.1373 | 0.335037 | standard |
| 7.31543 | 0.337117 | standard |
| 7.39498 | 0.0170841 | standard |
| 7.44993 | 0.0524893 | standard |
| 7.51968 | 0.316529 | standard |
| 7.53041 | 0.319826 | standard |
| 7.61236 | 0.416528 | standard |
| 7.68044 | 0.0498161 | standard |
| 7.69695 | 0.061842 | standard |
| 7.77903 | 0.0156011 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.66786 | standard |
| 7.13729 | 0.33513 | standard |
| 7.31538 | 0.33694 | standard |
| 7.46054 | 0.0597903 | standard |
| 7.52037 | 0.312411 | standard |
| 7.52657 | 0.310712 | standard |
| 7.61395 | 0.397257 | standard |
| 7.69034 | 0.0700664 | standard |
| 7.76598 | 0.017094 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.66631 | standard |
| 7.13729 | 0.333366 | standard |
| 7.31538 | 0.334068 | standard |
| 7.48998 | 0.0980331 | standard |
| 7.52325 | 0.31305 | standard |
| 7.57606 | 0.0627843 | standard |
| 7.61759 | 0.232051 | standard |
| 7.62547 | 0.246513 | standard |
| 7.66695 | 0.111267 | standard |
| 7.70805 | 0.0302241 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.665762 | standard |
| 7.13729 | 0.332979 | standard |
| 7.31543 | 0.33316 | standard |
| 7.49854 | 0.111778 | standard |
| 7.52127 | 0.329757 | standard |
| 7.58547 | 0.0667001 | standard |
| 7.61404 | 0.180615 | standard |
| 7.63391 | 0.148468 | standard |
| 7.66168 | 0.129511 | standard |
| 7.6897 | 0.0391521 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.664389 | standard |
| 7.13734 | 0.333382 | standard |
| 7.31538 | 0.333484 | standard |
| 7.50186 | 0.11645 | standard |
| 7.51971 | 0.348577 | standard |
| 7.59024 | 0.0754921 | standard |
| 7.61213 | 0.166031 | standard |
| 7.63847 | 0.126719 | standard |
| 7.65961 | 0.135501 | standard |
| 7.68092 | 0.044533 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.66642 | standard |
| 7.13729 | 0.333971 | standard |
| 7.31538 | 0.333423 | standard |
| 7.50379 | 0.118769 | standard |
| 7.51857 | 0.364988 | standard |
| 7.59322 | 0.0824294 | standard |
| 7.59508 | 0.0802214 | standard |
| 7.61086 | 0.161412 | standard |
| 7.64156 | 0.114884 | standard |
| 7.65868 | 0.135673 | standard |
| 7.67582 | 0.046798 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.666644 | standard |
| 7.13729 | 0.333038 | standard |
| 7.31538 | 0.332204 | standard |
| 7.50511 | 0.119283 | standard |
| 7.51754 | 0.382194 | standard |
| 7.59508 | 0.0863136 | standard |
| 7.59685 | 0.0843707 | standard |
| 7.61002 | 0.1572 | standard |
| 7.64376 | 0.106126 | standard |
| 7.65824 | 0.134901 | standard |
| 7.6723 | 0.0484457 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.66477 | standard |
| 7.13729 | 0.332388 | standard |
| 7.31543 | 0.332421 | standard |
| 7.50608 | 0.119334 | standard |
| 7.51702 | 0.394493 | standard |
| 7.59647 | 0.091489 | standard |
| 7.59793 | 0.0898643 | standard |
| 7.60922 | 0.153047 | standard |
| 7.6454 | 0.0976548 | standard |
| 7.65801 | 0.134701 | standard |
| 7.66966 | 0.0515036 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.666669 | standard |
| 7.13729 | 0.332806 | standard |
| 7.31543 | 0.332835 | standard |
| 7.50637 | 0.12 | standard |
| 7.51687 | 0.398096 | standard |
| 7.59698 | 0.0924442 | standard |
| 7.59843 | 0.0909046 | standard |
| 7.60894 | 0.149383 | standard |
| 7.64606 | 0.0935449 | standard |
| 7.65785 | 0.136054 | standard |
| 7.66874 | 0.0542614 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.666718 | standard |
| 7.13729 | 0.332741 | standard |
| 7.31543 | 0.332766 | standard |
| 7.50673 | 0.119709 | standard |
| 7.51673 | 0.397369 | standard |
| 7.59747 | 0.0941712 | standard |
| 7.59877 | 0.0924838 | standard |
| 7.60871 | 0.146269 | standard |
| 7.64669 | 0.0902227 | standard |
| 7.65773 | 0.135338 | standard |
| 7.66794 | 0.0558784 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.66665 | standard |
| 7.13729 | 0.332392 | standard |
| 7.31543 | 0.332412 | standard |
| 7.50723 | 0.119037 | standard |
| 7.51645 | 0.372773 | standard |
| 7.59827 | 0.0968201 | standard |
| 7.59935 | 0.0965129 | standard |
| 7.60799 | 0.140836 | standard |
| 7.64792 | 0.0857836 | standard |
| 7.65753 | 0.136742 | standard |
| 7.6666 | 0.0599945 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.665047 | standard |
| 7.13729 | 0.331593 | standard |
| 7.31543 | 0.331611 | standard |
| 7.50743 | 0.119859 | standard |
| 7.51628 | 0.358929 | standard |
| 7.59852 | 0.0950384 | standard |
| 7.5997 | 0.0956634 | standard |
| 7.60773 | 0.137911 | standard |
| 7.6484 | 0.0849223 | standard |
| 7.65738 | 0.138384 | standard |
| 7.6661 | 0.05997 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.665957 | standard |
| 7.13729 | 0.33175 | standard |
| 7.31543 | 0.331766 | standard |
| 7.50769 | 0.118449 | standard |
| 7.51611 | 0.342812 | standard |
| 7.59882 | 0.0964162 | standard |
| 7.5999 | 0.0970286 | standard |
| 7.60752 | 0.137013 | standard |
| 7.60813 | 0.136736 | standard |
| 7.64886 | 0.0851497 | standard |
| 7.65729 | 0.143091 | standard |
| 7.66561 | 0.0636503 | standard |
| 2.37779 | 1.0 | standard |
| 4.26758 | 0.666414 | standard |
| 7.13729 | 0.331629 | standard |
| 7.31543 | 0.331639 | standard |
| 7.50854 | 0.11843 | standard |
| 7.51533 | 0.215667 | standard |
| 7.51676 | 0.23017 | standard |
| 7.60007 | 0.100321 | standard |
| 7.60089 | 0.101237 | standard |
| 7.60662 | 0.129501 | standard |
| 7.60735 | 0.129093 | standard |
| 7.65068 | 0.0903557 | standard |
| 7.65708 | 0.15034 | standard |
| 7.66352 | 0.0685819 | standard |