{4-[5-(Trifluoromethyl)pyrid-2-yl]phenyl}methylamine
Simulation outputs:
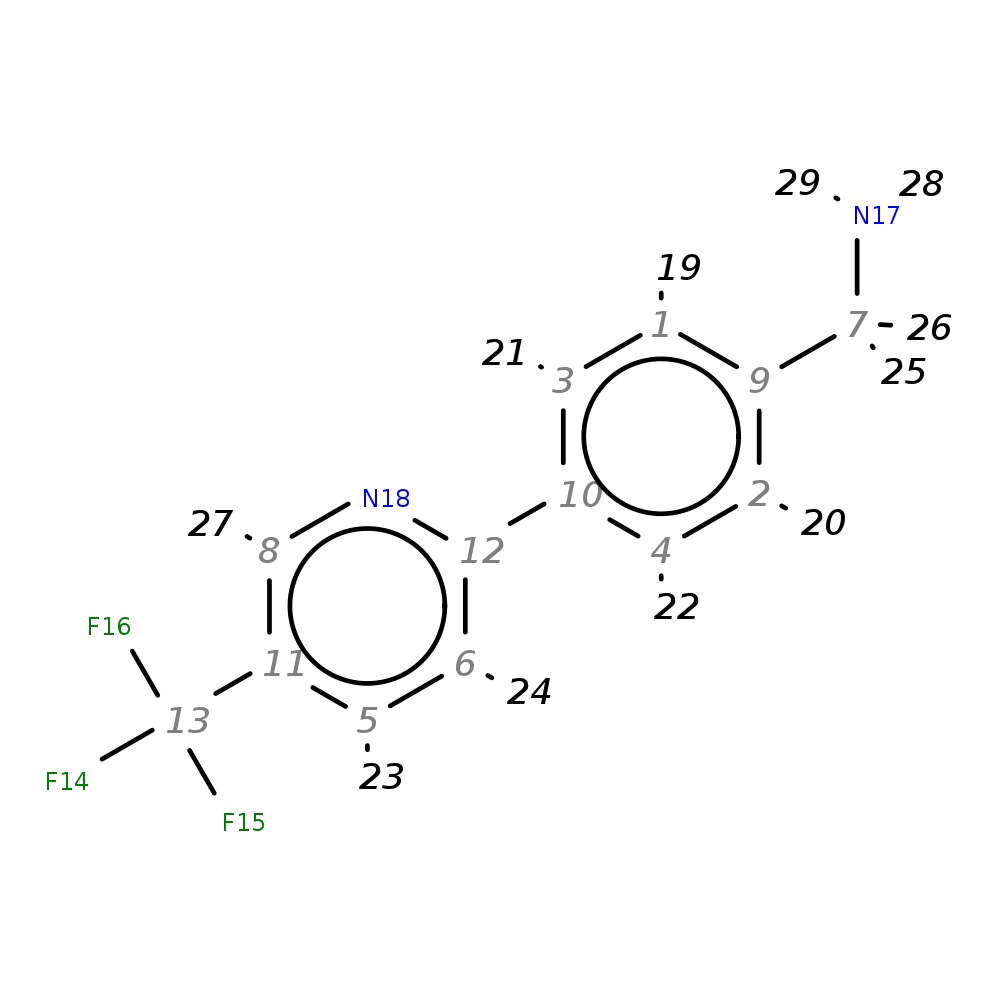
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02288 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C13H11F3N2/c14-13(15,16)11-5-6-12(18-8-11)10-3-1-9(7-17)2-4-10/h1-6,8H,7,17H2 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| {4-[5-(Trifluoromethyl)pyrid-2-yl]phenyl}methylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
|---|---|---|---|---|---|---|---|---|---|
| 19 | 7.633 | 1.5 | 7.782 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 7.633 | 0 | 7.782 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 8.024 | 1.5 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 8.024 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 8.278 | 8.607 | 0 | 0 | 2.204 |
| 24 | 0 | 0 | 0 | 0 | 0 | 8.056 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 4.279 | -14.0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.279 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.955 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.27869 | 1.0 | standard |
| 7.62679 | 0.417441 | standard |
| 7.63966 | 0.435968 | standard |
| 8.01738 | 0.436978 | standard |
| 8.03025 | 0.422166 | standard |
| 8.04837 | 0.243213 | standard |
| 8.06275 | 0.271161 | standard |
| 8.26977 | 0.159077 | standard |
| 8.27321 | 0.160626 | standard |
| 8.28414 | 0.142385 | standard |
| 8.28758 | 0.140606 | standard |
| 8.95356 | 0.293917 | standard |
| 8.957 | 0.293917 | standard |
| 4.27869 | 1.0 | standard |
| 7.51284 | 0.246669 | standard |
| 7.70854 | 0.625106 | standard |
| 7.94826 | 0.652784 | standard |
| 8.1228 | 0.617138 | standard |
| 8.18579 | 0.38822 | standard |
| 8.23345 | 0.313051 | standard |
| 8.40876 | 0.0559271 | standard |
| 8.45037 | 0.069388 | standard |
| 8.93542 | 0.311713 | standard |
| 8.97623 | 0.285763 | standard |
| 4.27869 | 1.0 | standard |
| 7.55826 | 0.298948 | standard |
| 7.6877 | 0.558276 | standard |
| 7.96842 | 0.664291 | standard |
| 8.10341 | 0.659879 | standard |
| 8.21188 | 0.251968 | standard |
| 8.24285 | 0.250421 | standard |
| 8.35556 | 0.074359 | standard |
| 8.38647 | 0.0848461 | standard |
| 8.94027 | 0.306975 | standard |
| 8.97095 | 0.287533 | standard |
| 4.27869 | 1.0 | standard |
| 7.57906 | 0.329148 | standard |
| 7.67583 | 0.525761 | standard |
| 7.98261 | 0.631488 | standard |
| 8.08158 | 0.453511 | standard |
| 8.09555 | 0.497673 | standard |
| 8.22459 | 0.223593 | standard |
| 8.24882 | 0.223475 | standard |
| 8.33216 | 0.0885461 | standard |
| 8.35644 | 0.096439 | standard |
| 8.94345 | 0.302706 | standard |
| 8.96751 | 0.290386 | standard |
| 4.27869 | 1.0 | standard |
| 7.58571 | 0.339909 | standard |
| 7.67162 | 0.515403 | standard |
| 7.98641 | 0.58776 | standard |
| 8.0726 | 0.416773 | standard |
| 8.09255 | 0.455727 | standard |
| 8.22935 | 0.214636 | standard |
| 8.25115 | 0.215496 | standard |
| 8.32492 | 0.0951823 | standard |
| 8.34678 | 0.0998309 | standard |
| 8.94461 | 0.300703 | standard |
| 8.96631 | 0.292109 | standard |
| 4.27869 | 1.0 | standard |
| 7.59092 | 0.348707 | standard |
| 7.66817 | 0.506195 | standard |
| 7.98942 | 0.556189 | standard |
| 8.06682 | 0.403468 | standard |
| 8.08984 | 0.420735 | standard |
| 8.23343 | 0.207749 | standard |
| 8.25313 | 0.209204 | standard |
| 8.31937 | 0.0999773 | standard |
| 8.33923 | 0.103911 | standard |
| 8.94555 | 0.299771 | standard |
| 8.96533 | 0.291849 | standard |
| 4.27869 | 1.0 | standard |
| 7.61311 | 0.387562 | standard |
| 7.65164 | 0.467943 | standard |
| 8.00542 | 0.476364 | standard |
| 8.03322 | 0.316746 | standard |
| 8.04384 | 0.42579 | standard |
| 8.07516 | 0.31486 | standard |
| 8.25385 | 0.178726 | standard |
| 8.26405 | 0.17916 | standard |
| 8.29685 | 0.125007 | standard |
| 8.30707 | 0.123787 | standard |
| 8.95018 | 0.294287 | standard |
| 8.96043 | 0.294207 | standard |
| 4.27869 | 1.0 | standard |
| 7.62001 | 0.39951 | standard |
| 7.64574 | 0.455146 | standard |
| 8.0113 | 0.459157 | standard |
| 8.03769 | 0.517288 | standard |
| 8.06919 | 0.29167 | standard |
| 8.26156 | 0.168612 | standard |
| 8.26844 | 0.169734 | standard |
| 8.2902 | 0.133474 | standard |
| 8.29708 | 0.131862 | standard |
| 8.9519 | 0.294427 | standard |
| 8.95871 | 0.294368 | standard |
| 4.27869 | 1.0 | standard |
| 7.62345 | 0.406789 | standard |
| 7.6427 | 0.447984 | standard |
| 8.01435 | 0.450187 | standard |
| 8.03361 | 0.419736 | standard |
| 8.04443 | 0.259227 | standard |
| 8.06602 | 0.281099 | standard |
| 8.26556 | 0.163442 | standard |
| 8.27078 | 0.164829 | standard |
| 8.28707 | 0.137612 | standard |
| 8.29229 | 0.135864 | standard |
| 8.95273 | 0.294581 | standard |
| 8.95783 | 0.294581 | standard |
| 4.27869 | 1.0 | standard |
| 7.62552 | 0.417738 | standard |
| 7.64087 | 0.439349 | standard |
| 8.01621 | 0.441452 | standard |
| 8.03163 | 0.421296 | standard |
| 8.04677 | 0.245151 | standard |
| 8.06404 | 0.275108 | standard |
| 8.26811 | 0.161383 | standard |
| 8.2722 | 0.16286 | standard |
| 8.28525 | 0.140174 | standard |
| 8.28945 | 0.138369 | standard |
| 8.95318 | 0.292926 | standard |
| 8.95737 | 0.292925 | standard |
| 4.27869 | 1.0 | standard |
| 7.62679 | 0.417442 | standard |
| 7.63966 | 0.435969 | standard |
| 8.01738 | 0.436979 | standard |
| 8.03025 | 0.422168 | standard |
| 8.04837 | 0.243213 | standard |
| 8.06275 | 0.271161 | standard |
| 8.26977 | 0.159078 | standard |
| 8.27321 | 0.160626 | standard |
| 8.28414 | 0.142386 | standard |
| 8.28758 | 0.140607 | standard |
| 8.95356 | 0.293918 | standard |
| 8.957 | 0.293918 | standard |
| 4.27869 | 1.0 | standard |
| 7.62778 | 0.421066 | standard |
| 7.63878 | 0.435053 | standard |
| 8.01827 | 0.43584 | standard |
| 8.02926 | 0.422761 | standard |
| 8.04946 | 0.243366 | standard |
| 8.06176 | 0.268429 | standard |
| 8.27095 | 0.157368 | standard |
| 8.27393 | 0.158996 | standard |
| 8.28323 | 0.143397 | standard |
| 8.28621 | 0.141568 | standard |
| 8.95385 | 0.295675 | standard |
| 8.95671 | 0.295674 | standard |
| 4.27869 | 1.0 | standard |
| 7.62817 | 0.426647 | standard |
| 7.63838 | 0.431054 | standard |
| 8.01867 | 0.431627 | standard |
| 8.02898 | 0.425309 | standard |
| 8.04986 | 0.243646 | standard |
| 8.06136 | 0.267328 | standard |
| 8.27143 | 0.156631 | standard |
| 8.27423 | 0.158275 | standard |
| 8.28293 | 0.143728 | standard |
| 8.28573 | 0.141897 | standard |
| 8.95394 | 0.295473 | standard |
| 8.95662 | 0.295473 | standard |
| 4.27869 | 1.0 | standard |
| 7.62847 | 0.426987 | standard |
| 7.63808 | 0.431159 | standard |
| 8.01895 | 0.42905 | standard |
| 8.02858 | 0.42764 | standard |
| 8.05025 | 0.243994 | standard |
| 8.06096 | 0.266433 | standard |
| 8.27193 | 0.157851 | standard |
| 8.27442 | 0.159429 | standard |
| 8.28263 | 0.144332 | standard |
| 8.28523 | 0.14251 | standard |
| 8.95403 | 0.296017 | standard |
| 8.95652 | 0.296017 | standard |
| 4.27869 | 1.0 | standard |
| 7.62906 | 0.428094 | standard |
| 7.6376 | 0.428535 | standard |
| 8.01955 | 0.432264 | standard |
| 8.02809 | 0.425738 | standard |
| 8.05085 | 0.244592 | standard |
| 8.06047 | 0.264816 | standard |
| 8.27261 | 0.155405 | standard |
| 8.27494 | 0.157123 | standard |
| 8.28212 | 0.145005 | standard |
| 8.28445 | 0.143127 | standard |
| 8.95412 | 0.292851 | standard |
| 8.95644 | 0.292851 | standard |
| 4.27869 | 1.0 | standard |
| 7.62925 | 0.425846 | standard |
| 7.63729 | 0.432846 | standard |
| 8.01974 | 0.429829 | standard |
| 8.02779 | 0.426668 | standard |
| 8.05115 | 0.245272 | standard |
| 8.06017 | 0.263934 | standard |
| 8.27291 | 0.156184 | standard |
| 8.27504 | 0.157569 | standard |
| 8.28192 | 0.144833 | standard |
| 8.28416 | 0.143221 | standard |
| 8.95422 | 0.29509 | standard |
| 8.95634 | 0.29509 | standard |
| 4.27869 | 1.0 | standard |
| 7.62945 | 0.427461 | standard |
| 7.6371 | 0.429114 | standard |
| 8.01994 | 0.429525 | standard |
| 8.0276 | 0.428141 | standard |
| 8.05135 | 0.248827 | standard |
| 8.05997 | 0.260099 | standard |
| 8.2732 | 0.153576 | standard |
| 8.27524 | 0.155255 | standard |
| 8.28182 | 0.148381 | standard |
| 8.28386 | 0.146617 | standard |
| 8.9542 | 0.291263 | standard |
| 8.95635 | 0.291263 | standard |
| 4.27869 | 1.0 | standard |
| 7.63033 | 0.424966 | standard |
| 7.63632 | 0.426365 | standard |
| 8.02083 | 0.430238 | standard |
| 8.02671 | 0.430029 | standard |
| 8.05244 | 0.249617 | standard |
| 8.05898 | 0.258493 | standard |
| 8.27437 | 0.152526 | standard |
| 8.27595 | 0.15423 | standard |
| 8.28101 | 0.14895 | standard |
| 8.28259 | 0.14718 | standard |
| 8.95449 | 0.294156 | standard |
| 8.95607 | 0.294156 | standard |