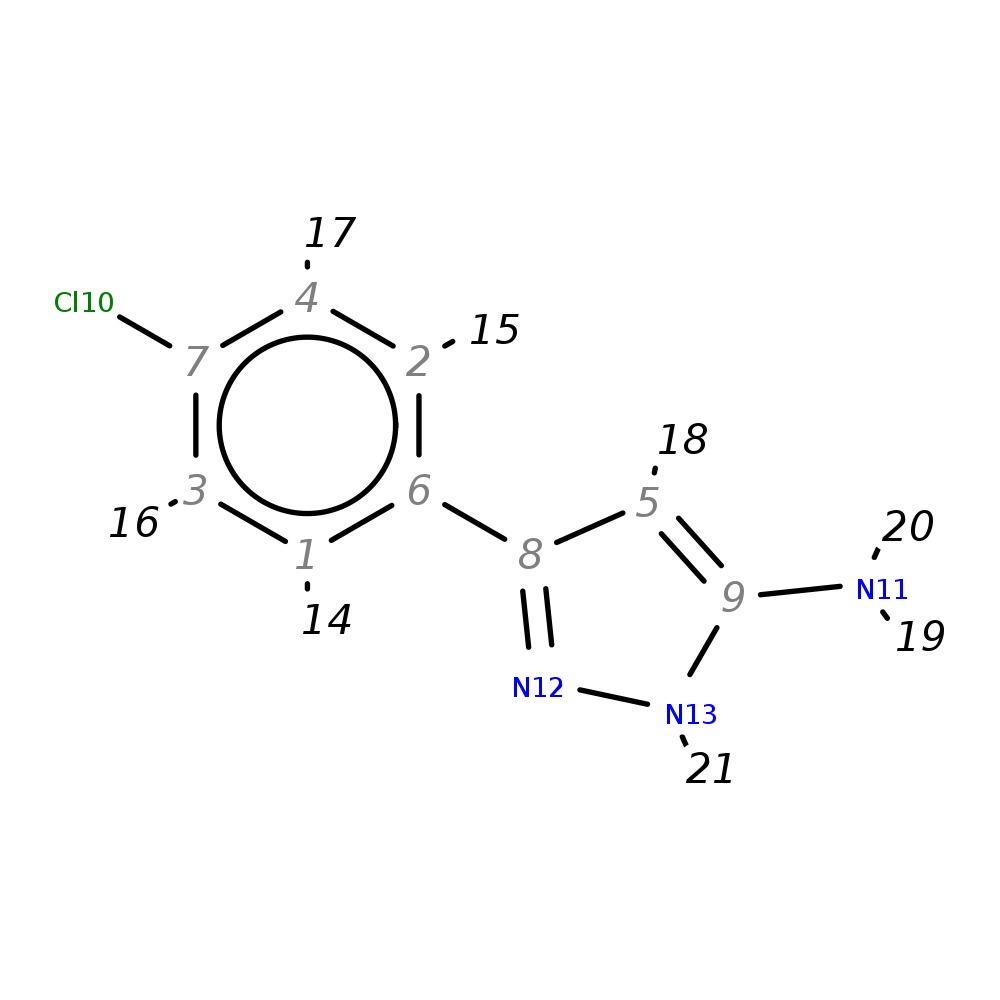
3-(4-Chlorophenyl)-1H-pyrazol-5-amine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03024 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H8ClN3/c10-7-3-1-6(2-4-7)8-5-9(11)13-12-8/h1-5H,(H3,11,12,13) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(4-Chlorophenyl)-1H-pyrazol-5-amine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|
| 14 | 7.503 | 1.5 | 8.79 | 0 | 0 |
| 15 | 0 | 7.503 | 0 | 8.79 | 0 |
| 16 | 0 | 0 | 7.655 | 1.5 | 0 |
| 17 | 0 | 0 | 0 | 7.655 | 0 |
| 18 | 0 | 0 | 0 | 0 | 6.082 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 6.0818 | 1.0 | standard |
| 7.49553 | 0.769312 | standard |
| 7.51003 | 0.92763 | standard |
| 7.64811 | 0.927509 | standard |
| 7.66267 | 0.771483 | standard |
| 6.0818 | 0.444506 | standard |
| 7.33299 | 0.0801493 | standard |
| 7.56474 | 1.0 | standard |
| 7.5934 | 0.999977 | standard |
| 7.82515 | 0.0800451 | standard |
| 6.0818 | 0.62015 | standard |
| 7.39869 | 0.173784 | standard |
| 7.54922 | 1.0 | standard |
| 7.60892 | 0.999998 | standard |
| 7.75946 | 0.173554 | standard |
| 6.0818 | 0.712094 | standard |
| 7.42964 | 0.261648 | standard |
| 7.54131 | 1.0 | standard |
| 7.61683 | 0.999997 | standard |
| 7.7285 | 0.261627 | standard |
| 6.0818 | 0.74444 | standard |
| 7.43942 | 0.301525 | standard |
| 7.53839 | 1.00001 | standard |
| 7.61982 | 0.999347 | standard |
| 7.71871 | 0.301426 | standard |
| 6.0818 | 0.771977 | standard |
| 7.44702 | 0.337942 | standard |
| 7.53579 | 1.0 | standard |
| 7.62235 | 0.999994 | standard |
| 7.71111 | 0.338171 | standard |
| 6.0818 | 0.92991 | standard |
| 7.47813 | 0.577131 | standard |
| 7.52197 | 0.998868 | standard |
| 7.6362 | 1.00008 | standard |
| 7.68001 | 0.577149 | standard |
| 6.0818 | 0.997503 | standard |
| 7.48716 | 0.693288 | standard |
| 7.51627 | 1.00011 | standard |
| 7.64186 | 1.00011 | standard |
| 7.67097 | 0.693286 | standard |
| 6.0818 | 1.0 | standard |
| 7.49143 | 0.732076 | standard |
| 7.51328 | 0.964546 | standard |
| 7.64485 | 0.964545 | standard |
| 7.66671 | 0.732075 | standard |
| 6.0818 | 1.0 | standard |
| 7.49389 | 0.756487 | standard |
| 7.51134 | 0.942065 | standard |
| 7.64683 | 0.943372 | standard |
| 7.66424 | 0.756525 | standard |
| 6.0818 | 1.0 | standard |
| 7.49553 | 0.769313 | standard |
| 7.51003 | 0.927634 | standard |
| 7.64811 | 0.927513 | standard |
| 7.66267 | 0.771484 | standard |
| 6.0818 | 1.0 | standard |
| 7.49664 | 0.781007 | standard |
| 7.50915 | 0.916206 | standard |
| 7.64909 | 0.916401 | standard |
| 7.66149 | 0.781238 | standard |
| 6.0818 | 1.0 | standard |
| 7.49711 | 0.78658 | standard |
| 7.50875 | 0.912894 | standard |
| 7.64938 | 0.912796 | standard |
| 7.6611 | 0.785002 | standard |
| 6.0818 | 1.0 | standard |
| 7.4975 | 0.791086 | standard |
| 7.50835 | 0.913752 | standard |
| 7.64978 | 0.907937 | standard |
| 7.66071 | 0.788274 | standard |
| 6.0818 | 1.0 | standard |
| 7.49812 | 0.79681 | standard |
| 7.50787 | 0.903574 | standard |
| 7.65037 | 0.902115 | standard |
| 7.66001 | 0.795665 | standard |
| 6.0818 | 1.0 | standard |
| 7.49842 | 0.799866 | standard |
| 7.50757 | 0.900928 | standard |
| 7.65057 | 0.902709 | standard |
| 7.65972 | 0.79988 | standard |
| 6.0818 | 1.0 | standard |
| 7.49861 | 0.799967 | standard |
| 7.50737 | 0.893952 | standard |
| 7.65076 | 0.895844 | standard |
| 7.65953 | 0.799982 | standard |
| 6.0818 | 1.0 | standard |
| 7.49969 | 0.811282 | standard |
| 7.50638 | 0.891901 | standard |
| 7.65175 | 0.882785 | standard |
| 7.65844 | 0.809265 | standard |