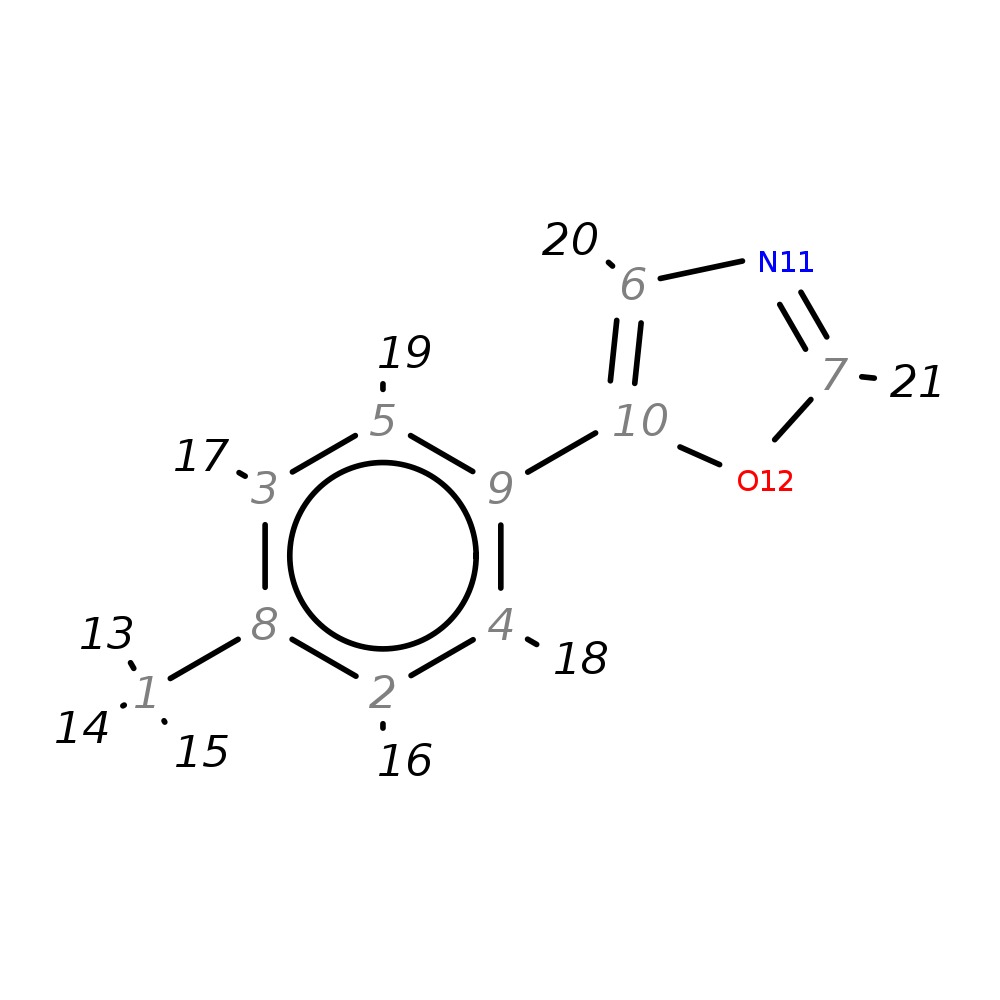
5-(4-Methylphenyl)-1,3-oxazole
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03404 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H9NO/c1-8-2-4-9(5-3-8)10-6-11-7-12-10/h2-7H,1H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 5-(4-Methylphenyl)-1,3-oxazole | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|
| 13 | 2.372 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.372 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.372 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 7.363 | 1.5 | 7.427 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 7.363 | 0 | 7.427 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 7.664 | 1.5 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 7.664 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.442 | 0.4 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.189 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.37223 | 1.0 | standard |
| 7.35652 | 0.278138 | standard |
| 7.36878 | 0.293864 | standard |
| 7.44174 | 0.323446 | standard |
| 7.65824 | 0.295201 | standard |
| 7.6705 | 0.276643 | standard |
| 8.18862 | 0.323115 | standard |
| 2.37223 | 1.0 | standard |
| 7.24298 | 0.148038 | standard |
| 7.43533 | 0.738768 | standard |
| 7.59618 | 0.455092 | standard |
| 7.78409 | 0.145677 | standard |
| 8.18867 | 0.322887 | standard |
| 2.37223 | 1.0 | standard |
| 7.28878 | 0.185989 | standard |
| 7.41488 | 0.48186 | standard |
| 7.43826 | 0.474358 | standard |
| 7.61433 | 0.396186 | standard |
| 7.73821 | 0.183369 | standard |
| 8.18862 | 0.321207 | standard |
| 2.37223 | 1.0 | standard |
| 7.30963 | 0.208561 | standard |
| 7.40244 | 0.396157 | standard |
| 7.44099 | 0.371005 | standard |
| 7.62491 | 0.367055 | standard |
| 7.7174 | 0.20647 | standard |
| 8.18862 | 0.32117 | standard |
| 2.37223 | 1.0 | standard |
| 7.3162 | 0.216525 | standard |
| 7.39846 | 0.377331 | standard |
| 7.44136 | 0.352892 | standard |
| 7.62867 | 0.358061 | standard |
| 7.71072 | 0.214769 | standard |
| 8.18862 | 0.321291 | standard |
| 2.37223 | 1.0 | standard |
| 7.32144 | 0.223224 | standard |
| 7.39525 | 0.364666 | standard |
| 7.44157 | 0.343443 | standard |
| 7.63176 | 0.350985 | standard |
| 7.70558 | 0.22143 | standard |
| 8.18862 | 0.321028 | standard |
| 2.37223 | 1.0 | standard |
| 7.34324 | 0.253611 | standard |
| 7.38 | 0.320882 | standard |
| 7.44174 | 0.32419 | standard |
| 7.64694 | 0.318539 | standard |
| 7.68374 | 0.252938 | standard |
| 8.18862 | 0.320722 | standard |
| 2.37223 | 1.0 | standard |
| 7.34994 | 0.26392 | standard |
| 7.37446 | 0.309041 | standard |
| 7.44174 | 0.320921 | standard |
| 7.65245 | 0.308487 | standard |
| 7.67697 | 0.263422 | standard |
| 8.18862 | 0.319517 | standard |
| 2.37223 | 1.0 | standard |
| 7.35328 | 0.270687 | standard |
| 7.37162 | 0.302959 | standard |
| 7.44174 | 0.32146 | standard |
| 7.65529 | 0.30159 | standard |
| 7.67374 | 0.269471 | standard |
| 8.18862 | 0.320697 | standard |
| 2.37223 | 1.0 | standard |
| 7.35524 | 0.273795 | standard |
| 7.36985 | 0.299319 | standard |
| 7.44174 | 0.321116 | standard |
| 7.65706 | 0.299042 | standard |
| 7.67178 | 0.272456 | standard |
| 8.18862 | 0.320636 | standard |
| 2.37223 | 1.0 | standard |
| 7.35652 | 0.278138 | standard |
| 7.36878 | 0.293828 | standard |
| 7.44174 | 0.323429 | standard |
| 7.65824 | 0.295202 | standard |
| 7.6705 | 0.276604 | standard |
| 8.18862 | 0.323098 | standard |
| 2.37223 | 1.0 | standard |
| 7.3574 | 0.28098 | standard |
| 7.36789 | 0.292477 | standard |
| 7.44174 | 0.31957 | standard |
| 7.65912 | 0.292476 | standard |
| 7.66962 | 0.279419 | standard |
| 8.18862 | 0.319328 | standard |
| 2.37223 | 1.0 | standard |
| 7.35779 | 0.283042 | standard |
| 7.3675 | 0.29008 | standard |
| 7.44174 | 0.317692 | standard |
| 7.65942 | 0.290243 | standard |
| 7.66922 | 0.282807 | standard |
| 8.18862 | 0.317482 | standard |
| 2.37223 | 1.0 | standard |
| 7.35809 | 0.282928 | standard |
| 7.3672 | 0.289914 | standard |
| 7.4417 | 0.320859 | standard |
| 7.65971 | 0.289801 | standard |
| 7.66893 | 0.281349 | standard |
| 8.18857 | 0.320595 | standard |
| 2.37223 | 1.0 | standard |
| 7.35857 | 0.283785 | standard |
| 7.36671 | 0.288692 | standard |
| 7.44174 | 0.323144 | standard |
| 7.6602 | 0.286442 | standard |
| 7.66833 | 0.28591 | standard |
| 8.18862 | 0.322999 | standard |
| 2.37223 | 1.0 | standard |
| 7.35877 | 0.284453 | standard |
| 7.36652 | 0.286884 | standard |
| 7.44174 | 0.321989 | standard |
| 7.66039 | 0.286895 | standard |
| 7.66814 | 0.284443 | standard |
| 8.18862 | 0.32186 | standard |
| 2.37223 | 1.0 | standard |
| 7.35896 | 0.283203 | standard |
| 7.36632 | 0.287703 | standard |
| 7.44174 | 0.320687 | standard |
| 7.66059 | 0.287608 | standard |
| 7.66795 | 0.285455 | standard |
| 8.18862 | 0.320569 | standard |
| 2.37223 | 1.0 | standard |
| 7.35985 | 0.283968 | standard |
| 7.36554 | 0.287291 | standard |
| 7.44174 | 0.312619 | standard |
| 7.66148 | 0.287428 | standard |
| 7.66706 | 0.287196 | standard |
| 8.18857 | 0.321289 | standard |