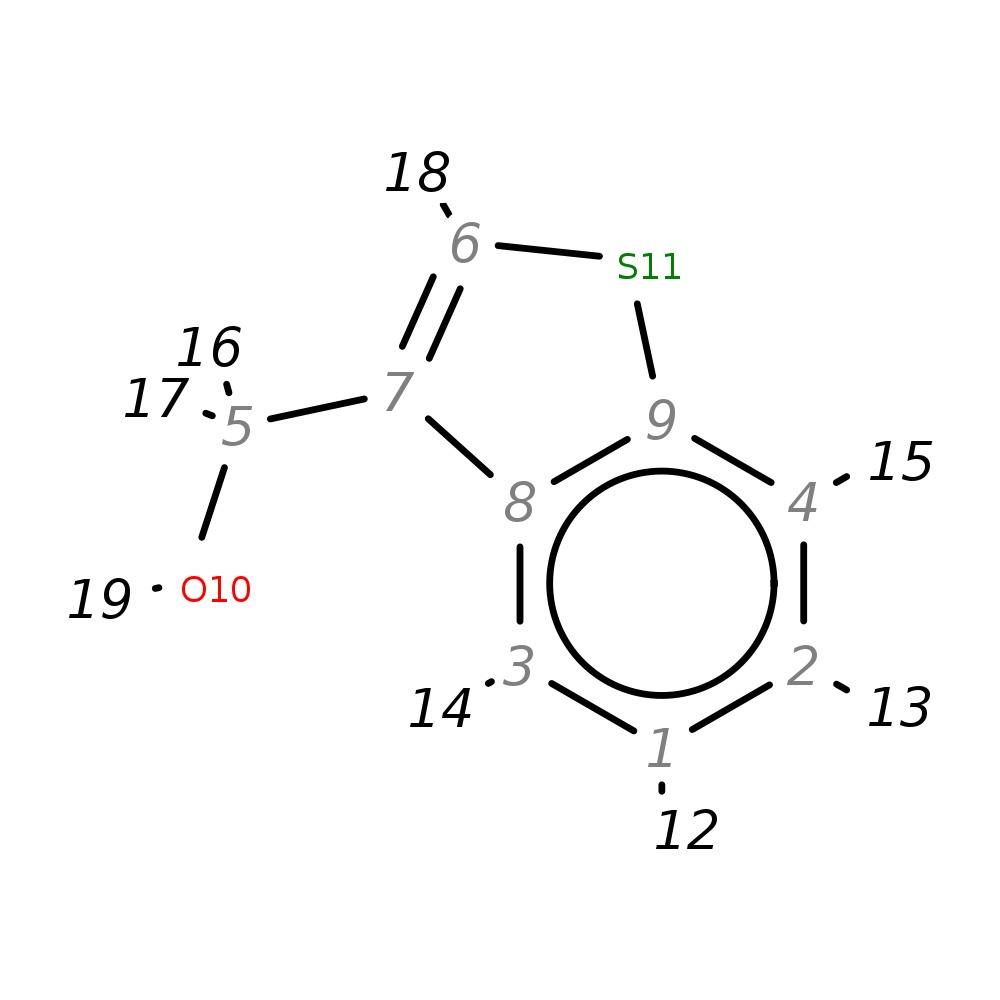
1-Benzothiophen-3-ylmethanol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.04389 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H8OS/c10-5-7-6-11-9-4-2-1-3-8(7)9/h1-4,6,10H,5H2 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 1-Benzothiophen-3-ylmethanol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|
| 12 | 7.467 | 8.324 | 8.528 | 0 | 0 | 0 | 0 |
| 13 | 0 | 7.505 | 0 | 8.528 | 0 | 0 | 0 |
| 14 | 0 | 0 | 8.014 | 0.5 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.932 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 4.909 | -14.0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 4.909 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 7.597 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.90912 | 1.0 | standard |
| 7.45183 | 0.0900815 | standard |
| 7.46568 | 0.234133 | standard |
| 7.47942 | 0.180923 | standard |
| 7.49265 | 0.175504 | standard |
| 7.50639 | 0.238069 | standard |
| 7.52023 | 0.0953976 | standard |
| 7.59717 | 0.500312 | standard |
| 7.92567 | 0.236013 | standard |
| 7.93942 | 0.230102 | standard |
| 8.00756 | 0.243694 | standard |
| 8.02129 | 0.22698 | standard |
| 4.90912 | 1.0 | standard |
| 7.22626 | 0.0291531 | standard |
| 7.35481 | 0.168438 | standard |
| 7.41333 | 0.17522 | standard |
| 7.59073 | 0.74899 | standard |
| 7.69819 | 0.180734 | standard |
| 7.85 | 0.241602 | standard |
| 7.92357 | 0.301074 | standard |
| 7.99568 | 0.210284 | standard |
| 8.06781 | 0.158335 | standard |
| 8.14235 | 0.106651 | standard |
| 4.90912 | 1.0 | standard |
| 7.30214 | 0.0288981 | standard |
| 7.32475 | 0.0273331 | standard |
| 7.40524 | 0.211651 | standard |
| 7.4295 | 0.183823 | standard |
| 7.45526 | 0.178506 | standard |
| 7.49803 | 0.218895 | standard |
| 7.54808 | 0.40829 | standard |
| 7.5969 | 0.558262 | standard |
| 7.76207 | 0.0399061 | standard |
| 7.7942 | 0.0513901 | standard |
| 7.87221 | 0.21653 | standard |
| 7.95282 | 0.250988 | standard |
| 7.98057 | 0.206446 | standard |
| 8.00859 | 0.191673 | standard |
| 8.07098 | 0.106833 | standard |
| 8.09399 | 0.123796 | standard |
| 8.17161 | 0.0198031 | standard |
| 4.90912 | 1.0 | standard |
| 7.34305 | 0.030344 | standard |
| 7.42932 | 0.232018 | standard |
| 7.47013 | 0.185066 | standard |
| 7.49116 | 0.204882 | standard |
| 7.53249 | 0.352107 | standard |
| 7.59707 | 0.549143 | standard |
| 7.79574 | 0.025232 | standard |
| 7.82698 | 0.039894 | standard |
| 7.88699 | 0.201325 | standard |
| 7.9669 | 0.252705 | standard |
| 7.99053 | 0.222531 | standard |
| 8.07103 | 0.133093 | standard |
| 8.13064 | 0.0201951 | standard |
| 4.90912 | 1.0 | standard |
| 7.35694 | 0.031263 | standard |
| 7.43776 | 0.239146 | standard |
| 7.47472 | 0.191549 | standard |
| 7.48923 | 0.205236 | standard |
| 7.52669 | 0.339554 | standard |
| 7.59734 | 0.543735 | standard |
| 7.80725 | 0.021368 | standard |
| 7.83828 | 0.03674 | standard |
| 7.89182 | 0.195808 | standard |
| 7.94394 | 0.139629 | standard |
| 7.97117 | 0.249699 | standard |
| 8.01215 | 0.137 | standard |
| 8.06388 | 0.135664 | standard |
| 8.11715 | 0.0200733 | standard |
| 8.14622 | 0.0105731 | standard |
| 4.90912 | 1.0 | standard |
| 7.3682 | 0.0323225 | standard |
| 7.44445 | 0.247593 | standard |
| 7.47909 | 0.201851 | standard |
| 7.48743 | 0.210235 | standard |
| 7.52143 | 0.332258 | standard |
| 7.59714 | 0.554086 | standard |
| 7.81662 | 0.0184331 | standard |
| 7.84727 | 0.0347205 | standard |
| 7.89603 | 0.189508 | standard |
| 7.93955 | 0.140465 | standard |
| 7.97677 | 0.27196 | standard |
| 8.01448 | 0.138258 | standard |
| 8.05802 | 0.138142 | standard |
| 8.10634 | 0.0203022 | standard |
| 8.1358 | 0.00979812 | standard |
| 4.90912 | 1.0 | standard |
| 7.4185 | 0.0463215 | standard |
| 7.45851 | 0.199659 | standard |
| 7.47132 | 0.163012 | standard |
| 7.48521 | 0.286086 | standard |
| 7.49925 | 0.185356 | standard |
| 7.51199 | 0.228127 | standard |
| 7.55201 | 0.0644108 | standard |
| 7.59717 | 0.502951 | standard |
| 7.8594 | 0.00687204 | standard |
| 7.88638 | 0.0232111 | standard |
| 7.91336 | 0.194089 | standard |
| 7.92711 | 0.0924609 | standard |
| 7.95384 | 0.150283 | standard |
| 7.99484 | 0.175402 | standard |
| 8.02151 | 0.0868581 | standard |
| 8.03527 | 0.167003 | standard |
| 8.06214 | 0.0176871 | standard |
| 8.08884 | 0.005016 | standard |
| 4.90912 | 1.0 | standard |
| 7.43528 | 0.0613283 | standard |
| 7.46261 | 0.204603 | standard |
| 7.48576 | 0.305947 | standard |
| 7.50886 | 0.220187 | standard |
| 7.5362 | 0.0734878 | standard |
| 7.59717 | 0.501177 | standard |
| 7.8959 | 0.0129041 | standard |
| 7.91926 | 0.207262 | standard |
| 7.94571 | 0.180246 | standard |
| 7.97485 | 0.0178311 | standard |
| 8.00201 | 0.202901 | standard |
| 8.0283 | 0.187243 | standard |
| 8.05163 | 0.010702 | standard |
| 4.90912 | 1.0 | standard |
| 7.44366 | 0.0735282 | standard |
| 7.4643 | 0.212368 | standard |
| 7.48589 | 0.270648 | standard |
| 7.50751 | 0.223419 | standard |
| 7.5282 | 0.0830495 | standard |
| 7.59717 | 0.50063 | standard |
| 7.8986 | 0.006541 | standard |
| 7.92247 | 0.219903 | standard |
| 7.9425 | 0.213781 | standard |
| 7.9684 | 0.00719025 | standard |
| 7.9789 | 0.00727656 | standard |
| 8.00476 | 0.234322 | standard |
| 8.02477 | 0.204779 | standard |
| 8.04858 | 0.00568802 | standard |
| 4.90912 | 1.0 | standard |
| 7.44863 | 0.081833 | standard |
| 7.46514 | 0.225309 | standard |
| 7.48139 | 0.190469 | standard |
| 7.49058 | 0.18308 | standard |
| 7.50681 | 0.230919 | standard |
| 7.52339 | 0.0902045 | standard |
| 7.59717 | 0.500394 | standard |
| 7.9244 | 0.230356 | standard |
| 7.94068 | 0.227189 | standard |
| 8.00638 | 0.244505 | standard |
| 8.0227 | 0.216502 | standard |
| 4.90912 | 1.0 | standard |
| 7.45183 | 0.0900805 | standard |
| 7.46568 | 0.234133 | standard |
| 7.47942 | 0.180925 | standard |
| 7.49265 | 0.175505 | standard |
| 7.50639 | 0.238069 | standard |
| 7.52023 | 0.0953977 | standard |
| 7.59717 | 0.500313 | standard |
| 7.92567 | 0.236014 | standard |
| 7.93942 | 0.230102 | standard |
| 8.00756 | 0.243695 | standard |
| 8.02129 | 0.226981 | standard |
| 4.90912 | 1.0 | standard |
| 7.45415 | 0.094875 | standard |
| 7.46605 | 0.239797 | standard |
| 7.47794 | 0.17328 | standard |
| 7.49423 | 0.170407 | standard |
| 7.50611 | 0.240171 | standard |
| 7.51801 | 0.099385 | standard |
| 7.59717 | 0.500253 | standard |
| 7.92665 | 0.237918 | standard |
| 7.9385 | 0.232409 | standard |
| 8.00841 | 0.243536 | standard |
| 8.02031 | 0.231453 | standard |
| 4.90912 | 1.0 | standard |
| 7.45505 | 0.0966687 | standard |
| 7.46616 | 0.241583 | standard |
| 7.47729 | 0.170926 | standard |
| 7.49487 | 0.168008 | standard |
| 7.506 | 0.24294 | standard |
| 7.51712 | 0.0994588 | standard |
| 7.59717 | 0.500243 | standard |
| 7.92703 | 0.236826 | standard |
| 7.93811 | 0.237164 | standard |
| 8.00881 | 0.2417 | standard |
| 8.01993 | 0.231922 | standard |
| 4.90912 | 1.0 | standard |
| 7.45584 | 0.0992955 | standard |
| 7.46626 | 0.242761 | standard |
| 7.4767 | 0.166125 | standard |
| 7.49551 | 0.165868 | standard |
| 7.50593 | 0.243892 | standard |
| 7.51632 | 0.100841 | standard |
| 7.59717 | 0.50019 | standard |
| 7.92733 | 0.239881 | standard |
| 7.93777 | 0.235975 | standard |
| 8.00912 | 0.240256 | standard |
| 8.01958 | 0.236217 | standard |
| 4.90912 | 1.0 | standard |
| 7.45717 | 0.102498 | standard |
| 7.46641 | 0.245578 | standard |
| 7.47571 | 0.163087 | standard |
| 7.4965 | 0.160802 | standard |
| 7.50578 | 0.244831 | standard |
| 7.51509 | 0.103804 | standard |
| 7.59717 | 0.50015 | standard |
| 7.92788 | 0.240816 | standard |
| 7.9372 | 0.235783 | standard |
| 8.00967 | 0.238469 | standard |
| 8.01899 | 0.238444 | standard |
| 4.90912 | 1.0 | standard |
| 7.45767 | 0.103601 | standard |
| 7.46651 | 0.246076 | standard |
| 7.47532 | 0.161014 | standard |
| 7.4969 | 0.158673 | standard |
| 7.50573 | 0.246784 | standard |
| 7.51449 | 0.104954 | standard |
| 7.59717 | 0.500134 | standard |
| 7.92811 | 0.236867 | standard |
| 7.93698 | 0.237408 | standard |
| 8.00989 | 0.239964 | standard |
| 8.01876 | 0.234647 | standard |
| 4.90912 | 1.0 | standard |
| 7.45817 | 0.104746 | standard |
| 7.46653 | 0.246809 | standard |
| 7.47492 | 0.159137 | standard |
| 7.49734 | 0.159259 | standard |
| 7.50571 | 0.24449 | standard |
| 7.51405 | 0.106192 | standard |
| 7.59717 | 0.500109 | standard |
| 7.92833 | 0.236969 | standard |
| 7.93675 | 0.239357 | standard |
| 8.01011 | 0.240515 | standard |
| 8.01854 | 0.235985 | standard |
| 4.90912 | 1.0 | standard |
| 7.4603 | 0.109987 | standard |
| 7.46673 | 0.248198 | standard |
| 7.47323 | 0.151196 | standard |
| 7.49903 | 0.151208 | standard |
| 7.50553 | 0.248216 | standard |
| 7.51197 | 0.110012 | standard |
| 7.59717 | 0.500057 | standard |
| 7.92925 | 0.237872 | standard |
| 7.93576 | 0.240974 | standard |
| 8.01108 | 0.237905 | standard |
| 8.01757 | 0.237202 | standard |