2-Methoxypyridin-3-amine
Simulation outputs:
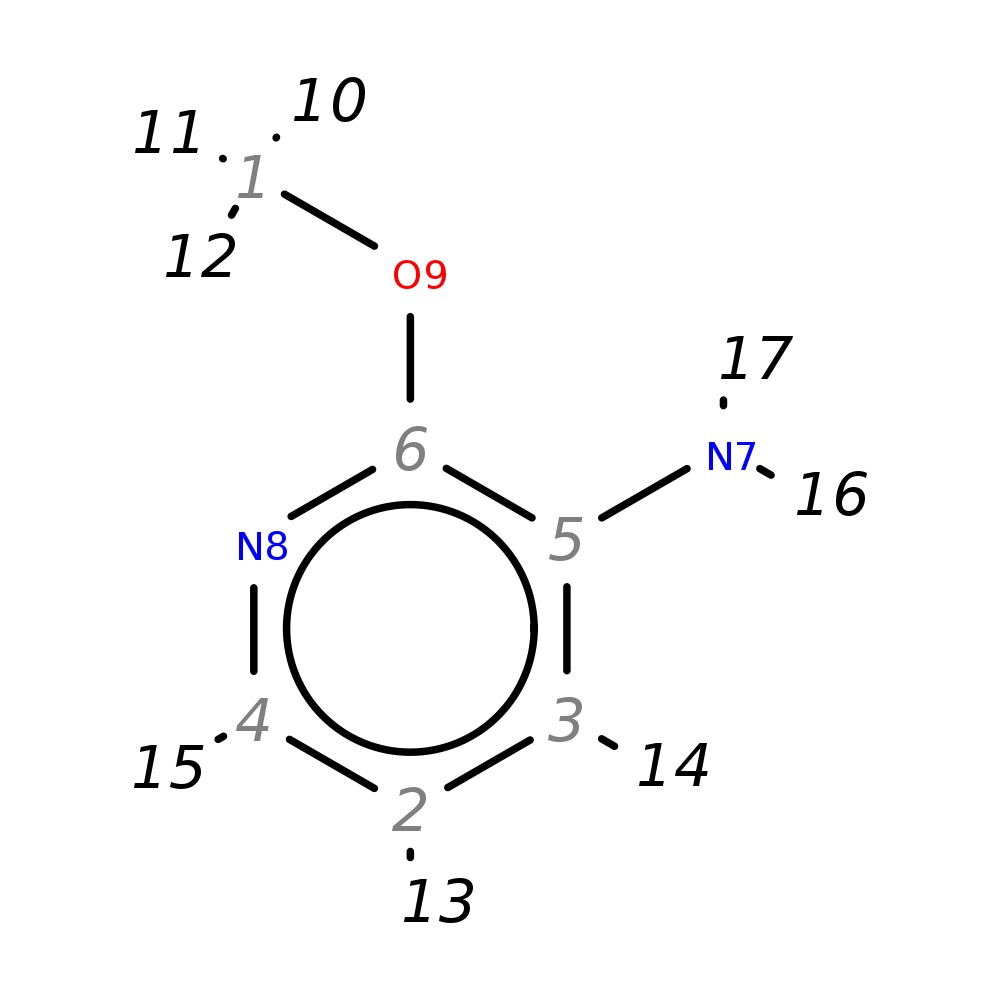
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02080 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H8N2O/c1-9-6-5(7)3-2-4-8-6/h2-4H,7H2,1H3 | |
| Note 1 | intensities strange |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-Methoxypyridin-3-amine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|
| 10 | 3.939 | -14.0 | -14.0 | 0 | 0 | 0 |
| 11 | 0 | 3.939 | -14.0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 3.939 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 6.889 | 7.856 | 5.54 |
| 14 | 0 | 0 | 0 | 0 | 7.166 | 1.464 |
| 15 | 0 | 0 | 0 | 0 | 0 | 7.541 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.93918 | 1.0 | standard |
| 6.8778 | 0.0866469 | standard |
| 6.88724 | 0.106346 | standard |
| 6.89067 | 0.111824 | standard |
| 6.90006 | 0.0932168 | standard |
| 7.16057 | 0.133181 | standard |
| 7.17269 | 0.122514 | standard |
| 7.53734 | 0.133058 | standard |
| 7.5453 | 0.132686 | standard |
| 3.93918 | 1.0 | standard |
| 6.69066 | 0.0408401 | standard |
| 6.8258 | 0.064747 | standard |
| 6.88615 | 0.124461 | standard |
| 7.01388 | 0.204532 | standard |
| 7.07758 | 0.191436 | standard |
| 7.11934 | 0.210914 | standard |
| 7.27194 | 0.069996 | standard |
| 7.46596 | 0.150261 | standard |
| 7.50527 | 0.136865 | standard |
| 7.58894 | 0.11144 | standard |
| 7.62777 | 0.0960031 | standard |
| 3.93918 | 1.0 | standard |
| 6.76227 | 0.0517041 | standard |
| 6.85402 | 0.0791461 | standard |
| 6.89218 | 0.121151 | standard |
| 6.98054 | 0.151901 | standard |
| 7.12634 | 0.181472 | standard |
| 7.23499 | 0.0785914 | standard |
| 7.49087 | 0.143106 | standard |
| 7.50994 | 0.138576 | standard |
| 7.57699 | 0.116517 | standard |
| 3.93918 | 1.0 | standard |
| 6.79651 | 0.0589711 | standard |
| 6.86624 | 0.0873071 | standard |
| 6.89372 | 0.119187 | standard |
| 6.96143 | 0.13419 | standard |
| 7.13261 | 0.168458 | standard |
| 7.21704 | 0.0875242 | standard |
| 7.50376 | 0.140919 | standard |
| 7.5148 | 0.139058 | standard |
| 7.56951 | 0.119628 | standard |
| 3.93918 | 1.0 | standard |
| 6.80756 | 0.061729 | standard |
| 6.86975 | 0.090116 | standard |
| 6.89391 | 0.11871 | standard |
| 6.95449 | 0.12898 | standard |
| 7.13517 | 0.164636 | standard |
| 7.21109 | 0.0917624 | standard |
| 7.50812 | 0.140265 | standard |
| 7.51713 | 0.139132 | standard |
| 7.56685 | 0.121495 | standard |
| 3.93918 | 1.0 | standard |
| 6.81635 | 0.0640653 | standard |
| 6.8725 | 0.092402 | standard |
| 6.894 | 0.118146 | standard |
| 6.94885 | 0.12474 | standard |
| 7.1375 | 0.160518 | standard |
| 7.20645 | 0.0948871 | standard |
| 7.51159 | 0.139352 | standard |
| 7.51916 | 0.1388 | standard |
| 7.56459 | 0.122583 | standard |
| 3.93918 | 1.0 | standard |
| 6.85418 | 0.0762204 | standard |
| 6.88249 | 0.101601 | standard |
| 6.89287 | 0.114894 | standard |
| 6.92083 | 0.106327 | standard |
| 7.14975 | 0.145426 | standard |
| 7.18585 | 0.111949 | standard |
| 7.52959 | 0.137764 | standard |
| 7.55341 | 0.129191 | standard |
| 3.93918 | 1.0 | standard |
| 6.86614 | 0.0816856 | standard |
| 6.88505 | 0.103676 | standard |
| 6.8919 | 0.11367 | standard |
| 6.91064 | 0.100145 | standard |
| 7.15495 | 0.139423 | standard |
| 7.17917 | 0.117091 | standard |
| 7.53347 | 0.135187 | standard |
| 7.54936 | 0.131594 | standard |
| 3.93918 | 1.0 | standard |
| 6.87207 | 0.0840584 | standard |
| 6.88618 | 0.104604 | standard |
| 6.89139 | 0.112997 | standard |
| 6.9054 | 0.0968812 | standard |
| 7.15764 | 0.13736 | standard |
| 7.176 | 0.12045 | standard |
| 7.53531 | 0.134505 | standard |
| 7.54734 | 0.133293 | standard |
| 3.93918 | 1.0 | standard |
| 6.87553 | 0.0855284 | standard |
| 6.8868 | 0.105323 | standard |
| 6.89094 | 0.112357 | standard |
| 6.90223 | 0.095088 | standard |
| 7.15943 | 0.134681 | standard |
| 7.17417 | 0.123117 | standard |
| 7.53652 | 0.132985 | standard |
| 7.54622 | 0.134815 | standard |
| 3.93918 | 1.0 | standard |
| 6.8778 | 0.086644 | standard |
| 6.88724 | 0.106364 | standard |
| 6.89067 | 0.111852 | standard |
| 6.90006 | 0.0939705 | standard |
| 7.16057 | 0.133193 | standard |
| 7.17272 | 0.122915 | standard |
| 7.53734 | 0.133083 | standard |
| 7.54532 | 0.13308 | standard |
| 3.93918 | 1.0 | standard |
| 6.87949 | 0.0872215 | standard |
| 6.88749 | 0.106115 | standard |
| 6.8905 | 0.110542 | standard |
| 6.89857 | 0.093354 | standard |
| 7.16133 | 0.13342 | standard |
| 7.17187 | 0.12476 | standard |
| 7.53778 | 0.1337 | standard |
| 7.54477 | 0.13402 | standard |
| 3.93918 | 1.0 | standard |
| 6.88008 | 0.0872663 | standard |
| 6.88769 | 0.107115 | standard |
| 6.89041 | 0.11155 | standard |
| 6.89788 | 0.0932843 | standard |
| 7.16171 | 0.132127 | standard |
| 7.17148 | 0.123886 | standard |
| 7.53805 | 0.132917 | standard |
| 7.54451 | 0.132901 | standard |
| 3.93918 | 1.0 | standard |
| 6.88067 | 0.0875477 | standard |
| 6.88777 | 0.107382 | standard |
| 6.89031 | 0.111478 | standard |
| 6.89739 | 0.0930374 | standard |
| 7.16208 | 0.131179 | standard |
| 7.17111 | 0.123531 | standard |
| 7.5383 | 0.132404 | standard |
| 7.54425 | 0.132404 | standard |
| 3.93918 | 1.0 | standard |
| 6.88166 | 0.0882119 | standard |
| 6.88794 | 0.10778 | standard |
| 6.89023 | 0.110461 | standard |
| 6.8965 | 0.0916678 | standard |
| 7.16254 | 0.129383 | standard |
| 7.17055 | 0.124002 | standard |
| 7.53866 | 0.13206 | standard |
| 7.5439 | 0.131622 | standard |
| 3.93918 | 1.0 | standard |
| 6.88206 | 0.0887049 | standard |
| 6.88793 | 0.10733 | standard |
| 6.89014 | 0.109419 | standard |
| 6.8961 | 0.091873 | standard |
| 7.16277 | 0.129388 | standard |
| 7.17034 | 0.12499 | standard |
| 7.53876 | 0.131989 | standard |
| 7.54379 | 0.132305 | standard |
| 3.93918 | 1.0 | standard |
| 6.88235 | 0.0891791 | standard |
| 6.88802 | 0.107013 | standard |
| 6.89015 | 0.109171 | standard |
| 6.8957 | 0.091576 | standard |
| 7.16276 | 0.134049 | standard |
| 7.17022 | 0.126042 | standard |
| 7.53887 | 0.132981 | standard |
| 7.54368 | 0.132785 | standard |
| 3.93918 | 1.0 | standard |
| 6.88394 | 0.0897767 | standard |
| 6.88829 | 0.108787 | standard |
| 6.88987 | 0.109476 | standard |
| 6.89422 | 0.0905817 | standard |
| 7.16336 | 0.125823 | standard |
| 7.1638 | 0.126545 | standard |
| 7.16941 | 0.130153 | standard |
| 7.53949 | 0.131073 | standard |
| 7.54307 | 0.131017 | standard |