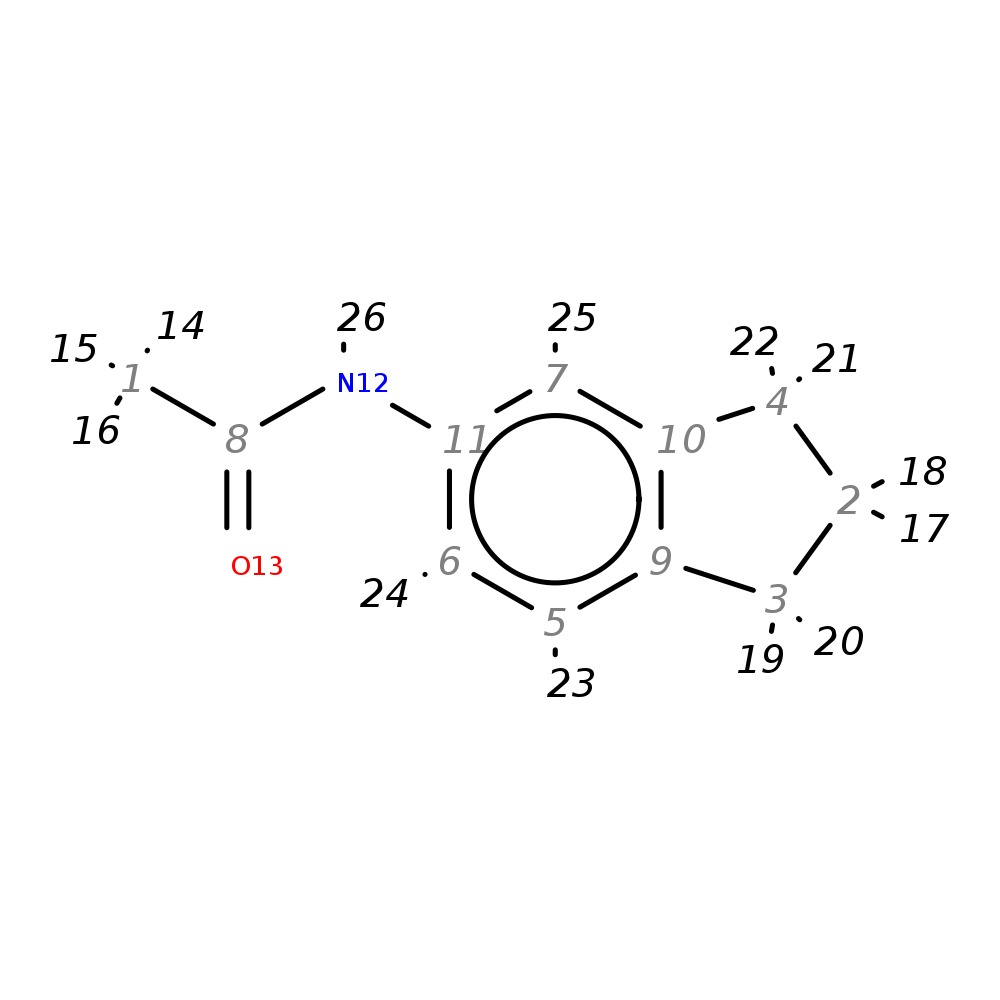
N1-(2,3-dihydro-1H-inden-5-yl)acetamide
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02044 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H13NO/c1-8(13)12-11-6-5-9-3-2-4-10(9)7-11/h5-7H,2-4H2,1H3,(H,12,13) | |
| Note 1 | 19,20?21,22 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N1-(2,3-dihydro-1H-inden-5-yl)acetamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 2.272 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.272 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.272 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 2.066 | -14.0 | 7.992 | 7.992 | 7.992 | 7.992 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 2.066 | 7.992 | 7.992 | 7.992 | 7.992 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 2.913 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 2.913 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.918 | -14.0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.918 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.341 | 8.108 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.4 | 1.539 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.517 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.03949 | 0.0437739 | standard |
| 2.05276 | 0.170579 | standard |
| 2.06605 | 0.253538 | standard |
| 2.07941 | 0.170192 | standard |
| 2.0927 | 0.0463666 | standard |
| 2.27166 | 1.0 | standard |
| 2.89997 | 0.193562 | standard |
| 2.90484 | 0.203722 | standard |
| 2.91334 | 0.380361 | standard |
| 2.9181 | 0.380001 | standard |
| 2.92661 | 0.201498 | standard |
| 2.93138 | 0.191465 | standard |
| 7.33393 | 0.132405 | standard |
| 7.34747 | 0.205861 | standard |
| 7.39305 | 0.136527 | standard |
| 7.39504 | 0.1372 | standard |
| 7.40665 | 0.0897271 | standard |
| 7.40848 | 0.089695 | standard |
| 7.51565 | 0.221672 | standard |
| 7.51755 | 0.220243 | standard |
| 1.59709 | 0.012053 | standard |
| 1.67699 | 0.0164191 | standard |
| 1.73362 | 0.0189911 | standard |
| 1.82398 | 0.0599981 | standard |
| 1.87447 | 0.0646711 | standard |
| 2.00628 | 0.0957711 | standard |
| 2.06223 | 0.142585 | standard |
| 2.27069 | 1.0 | standard |
| 2.41463 | 0.0953373 | standard |
| 2.75633 | 0.310082 | standard |
| 2.92006 | 0.455431 | standard |
| 3.10031 | 0.116801 | standard |
| 3.21298 | 0.0371961 | standard |
| 7.37091 | 0.528132 | standard |
| 7.5101 | 0.257784 | standard |
| 1.76675 | 0.0165471 | standard |
| 1.80305 | 0.0193671 | standard |
| 1.9149 | 0.080182 | standard |
| 1.93006 | 0.076258 | standard |
| 2.05533 | 0.15842 | standard |
| 2.19181 | 0.179237 | standard |
| 2.27182 | 1.0 | standard |
| 2.79863 | 0.311156 | standard |
| 2.91983 | 0.497659 | standard |
| 3.04569 | 0.154357 | standard |
| 7.23921 | 0.020422 | standard |
| 7.36992 | 0.483368 | standard |
| 7.51583 | 0.274605 | standard |
| 1.84803 | 0.019858 | standard |
| 1.86722 | 0.0216711 | standard |
| 1.95778 | 0.0959344 | standard |
| 2.05882 | 0.180876 | standard |
| 2.15877 | 0.156417 | standard |
| 2.2715 | 1.00001 | standard |
| 2.82414 | 0.311693 | standard |
| 2.91949 | 0.528489 | standard |
| 3.01645 | 0.18406 | standard |
| 7.26341 | 0.028956 | standard |
| 7.36655 | 0.430336 | standard |
| 7.51437 | 0.249249 | standard |
| 1.87451 | 0.021732 | standard |
| 1.88872 | 0.023217 | standard |
| 1.97114 | 0.104475 | standard |
| 2.06019 | 0.190462 | standard |
| 2.14886 | 0.157226 | standard |
| 2.27164 | 1.0 | standard |
| 2.83325 | 0.317713 | standard |
| 2.91913 | 0.552947 | standard |
| 3.006 | 0.199616 | standard |
| 7.27305 | 0.0333911 | standard |
| 7.36473 | 0.400028 | standard |
| 7.47859 | 0.0596571 | standard |
| 7.51414 | 0.249712 | standard |
| 1.89569 | 0.0233361 | standard |
| 1.90591 | 0.0244944 | standard |
| 1.98134 | 0.110603 | standard |
| 2.06127 | 0.198789 | standard |
| 2.14085 | 0.157708 | standard |
| 2.22595 | 0.087554 | standard |
| 2.27165 | 1.0 | standard |
| 2.84082 | 0.319049 | standard |
| 2.91874 | 0.565246 | standard |
| 2.9973 | 0.210939 | standard |
| 7.28094 | 0.0376121 | standard |
| 7.36261 | 0.369295 | standard |
| 7.46694 | 0.050877 | standard |
| 7.51397 | 0.247935 | standard |
| 1.98481 | 0.034409 | standard |
| 2.02472 | 0.142684 | standard |
| 2.06454 | 0.233627 | standard |
| 2.10435 | 0.171483 | standard |
| 2.14409 | 0.0498702 | standard |
| 2.27166 | 1.0 | standard |
| 2.87695 | 0.290563 | standard |
| 2.91652 | 0.540873 | standard |
| 2.95605 | 0.241687 | standard |
| 7.31495 | 0.0757327 | standard |
| 7.35542 | 0.266324 | standard |
| 7.38369 | 0.18649 | standard |
| 7.38849 | 0.188585 | standard |
| 7.4293 | 0.0589552 | standard |
| 7.51417 | 0.231758 | standard |
| 7.51858 | 0.220654 | standard |
| 2.01229 | 0.038451 | standard |
| 2.03885 | 0.156142 | standard |
| 2.06547 | 0.24651 | standard |
| 2.09208 | 0.174317 | standard |
| 2.11862 | 0.0488383 | standard |
| 2.27166 | 1.0 | standard |
| 2.88767 | 0.243031 | standard |
| 2.89128 | 0.246834 | standard |
| 2.91436 | 0.456124 | standard |
| 2.91767 | 0.455531 | standard |
| 2.94076 | 0.22062 | standard |
| 2.94433 | 0.216582 | standard |
| 7.32502 | 0.100517 | standard |
| 7.35203 | 0.238587 | standard |
| 7.38769 | 0.160611 | standard |
| 7.39134 | 0.163346 | standard |
| 7.41495 | 0.0681892 | standard |
| 7.41837 | 0.0721744 | standard |
| 7.5148 | 0.22692 | standard |
| 7.51832 | 0.21908 | standard |
| 2.02591 | 0.0404661 | standard |
| 2.04585 | 0.164456 | standard |
| 2.06581 | 0.251064 | standard |
| 2.08577 | 0.172163 | standard |
| 2.10574 | 0.0485576 | standard |
| 2.27166 | 1.0 | standard |
| 2.89365 | 0.217548 | standard |
| 2.89812 | 0.223058 | standard |
| 2.91366 | 0.414411 | standard |
| 2.91803 | 0.414197 | standard |
| 2.93352 | 0.208671 | standard |
| 2.938 | 0.203194 | standard |
| 7.32962 | 0.115619 | standard |
| 7.34995 | 0.223285 | standard |
| 7.3903 | 0.150743 | standard |
| 7.39296 | 0.152375 | standard |
| 7.41051 | 0.0771287 | standard |
| 7.41334 | 0.0793849 | standard |
| 7.5152 | 0.224992 | standard |
| 7.51793 | 0.220123 | standard |
| 2.03404 | 0.0432385 | standard |
| 2.04999 | 0.169768 | standard |
| 2.06599 | 0.25319 | standard |
| 2.08198 | 0.169488 | standard |
| 2.09795 | 0.0463592 | standard |
| 2.27166 | 1.0 | standard |
| 2.89741 | 0.200554 | standard |
| 2.90216 | 0.208376 | standard |
| 2.91344 | 0.392793 | standard |
| 2.9181 | 0.392505 | standard |
| 2.92935 | 0.205035 | standard |
| 2.93405 | 0.197727 | standard |
| 7.3323 | 0.125523 | standard |
| 7.34846 | 0.213215 | standard |
| 7.39196 | 0.143293 | standard |
| 7.39415 | 0.14425 | standard |
| 7.40809 | 0.083716 | standard |
| 7.41046 | 0.0850689 | standard |
| 7.51545 | 0.222753 | standard |
| 7.51772 | 0.219979 | standard |
| 2.03949 | 0.0437227 | standard |
| 2.05276 | 0.170379 | standard |
| 2.06605 | 0.253571 | standard |
| 2.07941 | 0.169993 | standard |
| 2.0927 | 0.0463126 | standard |
| 2.27166 | 1.0 | standard |
| 2.89997 | 0.193333 | standard |
| 2.90484 | 0.203483 | standard |
| 2.91334 | 0.379944 | standard |
| 2.9181 | 0.379884 | standard |
| 2.92661 | 0.201269 | standard |
| 2.93138 | 0.19124 | standard |
| 7.33393 | 0.132222 | standard |
| 7.34747 | 0.205396 | standard |
| 7.39305 | 0.1365 | standard |
| 7.39504 | 0.137185 | standard |
| 7.40665 | 0.0894516 | standard |
| 7.40848 | 0.0893568 | standard |
| 7.51565 | 0.221318 | standard |
| 7.51755 | 0.219801 | standard |
| 2.04329 | 0.0450762 | standard |
| 2.05472 | 0.170708 | standard |
| 2.06613 | 0.254071 | standard |
| 2.07756 | 0.170337 | standard |
| 2.08899 | 0.0450893 | standard |
| 2.27166 | 1.0 | standard |
| 2.90183 | 0.187495 | standard |
| 2.9067 | 0.201235 | standard |
| 2.91327 | 0.373039 | standard |
| 2.91808 | 0.372852 | standard |
| 2.92465 | 0.201321 | standard |
| 2.92951 | 0.187679 | standard |
| 7.33512 | 0.137465 | standard |
| 7.34672 | 0.200396 | standard |
| 7.394 | 0.133573 | standard |
| 7.39567 | 0.134057 | standard |
| 7.40559 | 0.0923813 | standard |
| 7.40725 | 0.0916368 | standard |
| 7.51582 | 0.222476 | standard |
| 7.51739 | 0.221855 | standard |
| 2.04488 | 0.0450465 | standard |
| 2.05551 | 0.170679 | standard |
| 2.06614 | 0.254734 | standard |
| 2.07681 | 0.170575 | standard |
| 2.08745 | 0.0453369 | standard |
| 2.27166 | 1.0 | standard |
| 2.90258 | 0.186164 | standard |
| 2.90748 | 0.201045 | standard |
| 2.91326 | 0.370791 | standard |
| 2.91809 | 0.370856 | standard |
| 2.92384 | 0.201671 | standard |
| 2.92873 | 0.185798 | standard |
| 7.33562 | 0.139528 | standard |
| 7.34642 | 0.19869 | standard |
| 7.3944 | 0.133461 | standard |
| 7.39586 | 0.134022 | standard |
| 7.4052 | 0.0947661 | standard |
| 7.40666 | 0.0942223 | standard |
| 7.51577 | 0.21863 | standard |
| 7.51739 | 0.21836 | standard |
| 2.04622 | 0.0453298 | standard |
| 2.05621 | 0.170732 | standard |
| 2.06614 | 0.254952 | standard |
| 2.07616 | 0.171355 | standard |
| 2.08617 | 0.0453208 | standard |
| 2.27166 | 1.0 | standard |
| 2.90325 | 0.185368 | standard |
| 2.90811 | 0.202216 | standard |
| 2.91326 | 0.369195 | standard |
| 2.91809 | 0.369486 | standard |
| 2.92314 | 0.202642 | standard |
| 2.9281 | 0.184973 | standard |
| 7.33602 | 0.141402 | standard |
| 7.34613 | 0.19748 | standard |
| 7.39464 | 0.129501 | standard |
| 7.39621 | 0.129864 | standard |
| 7.40488 | 0.0955307 | standard |
| 7.40629 | 0.0949495 | standard |
| 7.51588 | 0.221967 | standard |
| 7.51729 | 0.221966 | standard |
| 2.04845 | 0.0454891 | standard |
| 2.05732 | 0.171218 | standard |
| 2.06622 | 0.255078 | standard |
| 2.07507 | 0.171242 | standard |
| 2.08399 | 0.0452247 | standard |
| 2.27166 | 1.0 | standard |
| 2.90433 | 0.182891 | standard |
| 2.90921 | 0.205031 | standard |
| 2.91323 | 0.367145 | standard |
| 2.9181 | 0.367474 | standard |
| 2.92203 | 0.206036 | standard |
| 2.92691 | 0.18301 | standard |
| 7.33661 | 0.144308 | standard |
| 7.34563 | 0.19427 | standard |
| 7.39524 | 0.129588 | standard |
| 7.39651 | 0.129981 | standard |
| 7.40425 | 0.0972211 | standard |
| 7.40552 | 0.0965935 | standard |
| 7.51595 | 0.221243 | standard |
| 7.51721 | 0.221243 | standard |
| 2.04939 | 0.0452268 | standard |
| 2.05781 | 0.171296 | standard |
| 2.06623 | 0.255362 | standard |
| 2.0746 | 0.170961 | standard |
| 2.08299 | 0.0452459 | standard |
| 2.27166 | 1.0 | standard |
| 2.90483 | 0.182174 | standard |
| 2.90972 | 0.208538 | standard |
| 2.91319 | 0.367766 | standard |
| 2.91811 | 0.367321 | standard |
| 2.92153 | 0.209092 | standard |
| 2.92652 | 0.181807 | standard |
| 7.33691 | 0.145597 | standard |
| 7.34543 | 0.192976 | standard |
| 7.39553 | 0.129046 | standard |
| 7.39671 | 0.129465 | standard |
| 7.40405 | 0.0983158 | standard |
| 7.40523 | 0.097678 | standard |
| 7.51591 | 0.216648 | standard |
| 7.51725 | 0.216647 | standard |
| 2.05023 | 0.0454644 | standard |
| 2.05821 | 0.171053 | standard |
| 2.06623 | 0.255043 | standard |
| 2.0742 | 0.171594 | standard |
| 2.0822 | 0.0451559 | standard |
| 2.27166 | 1.0 | standard |
| 2.90522 | 0.181169 | standard |
| 2.91013 | 0.211496 | standard |
| 2.91319 | 0.368284 | standard |
| 2.91811 | 0.367843 | standard |
| 2.92112 | 0.212532 | standard |
| 2.92603 | 0.181249 | standard |
| 7.33711 | 0.146727 | standard |
| 7.34523 | 0.191746 | standard |
| 7.39573 | 0.128913 | standard |
| 7.39682 | 0.129332 | standard |
| 7.40385 | 0.0995308 | standard |
| 7.40494 | 0.0988954 | standard |
| 7.51604 | 0.222972 | standard |
| 7.51712 | 0.222951 | standard |
| 2.054 | 0.0454286 | standard |
| 2.06009 | 0.171902 | standard |
| 2.06624 | 0.255257 | standard |
| 2.07239 | 0.171893 | standard |
| 2.07853 | 0.0455597 | standard |
| 2.27166 | 1.0 | standard |
| 2.90705 | 0.178481 | standard |
| 2.91315 | 0.392322 | standard |
| 2.9181 | 0.392122 | standard |
| 2.9242 | 0.17848 | standard |
| 7.3382 | 0.151828 | standard |
| 7.34444 | 0.186579 | standard |
| 7.39666 | 0.123505 | standard |
| 7.39757 | 0.124095 | standard |
| 7.4029 | 0.101476 | standard |
| 7.40381 | 0.100738 | standard |
| 7.51619 | 0.223311 | standard |
| 7.51701 | 0.222737 | standard |