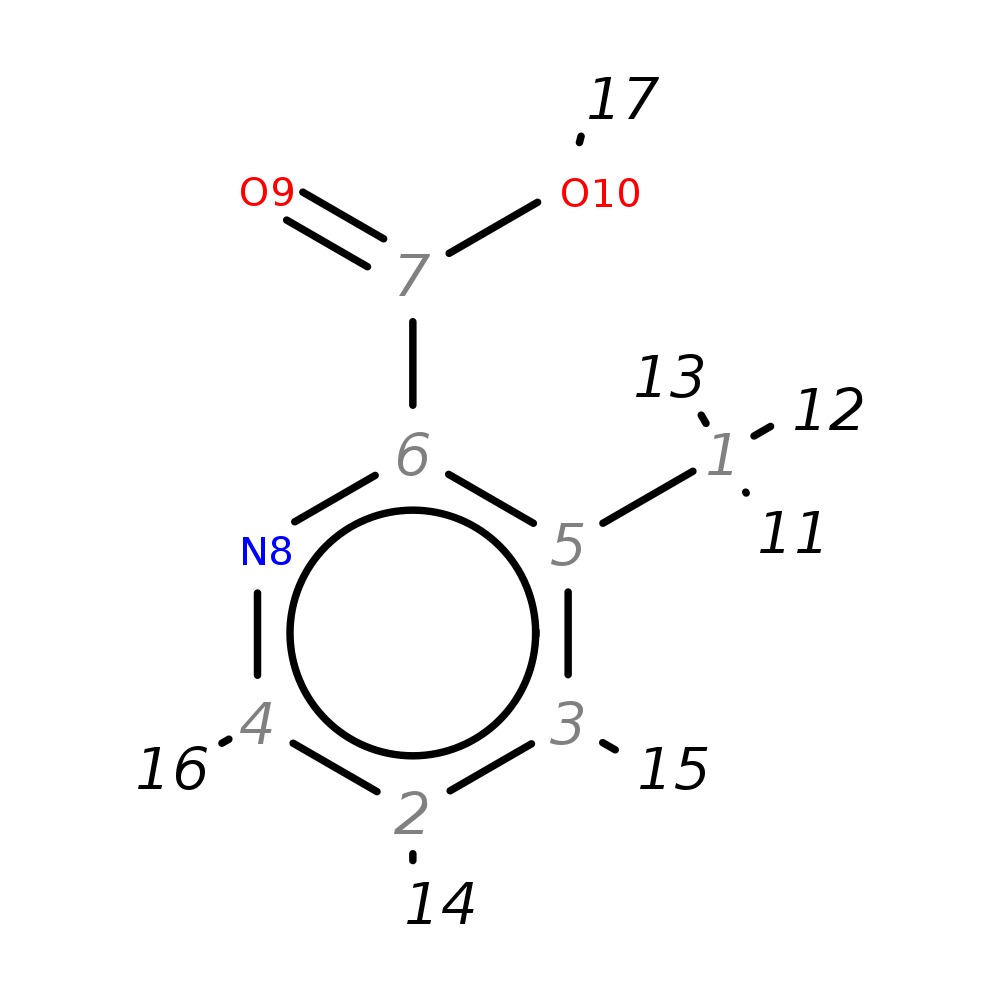
3-Methylpicolinic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03104 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H7NO2/c1-5-3-2-4-8-6(5)7(9)10/h2-4H,1H3,(H,9,10) | |
| Note 1 | 15?16 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Methylpicolinic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|
| 11 | 2.325 | -14.0 | -14.0 | 0 | 0 | 0 |
| 12 | 0 | 2.325 | -14.0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 2.325 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 7.323 | 7.125 | 4.704 |
| 15 | 0 | 0 | 0 | 0 | 7.721 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 8.249 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.32462 | 1.0 | standard |
| 7.32271 | 0.208994 | standard |
| 7.71619 | 0.213989 | standard |
| 7.72622 | 0.209382 | standard |
| 8.24934 | 0.253139 | standard |
| 2.32462 | 1.0 | standard |
| 7.43277 | 0.259764 | standard |
| 7.65038 | 0.311017 | standard |
| 8.22051 | 0.278039 | standard |
| 2.32462 | 1.0 | standard |
| 7.33576 | 0.222981 | standard |
| 7.67429 | 0.265856 | standard |
| 8.23221 | 0.266893 | standard |
| 2.32462 | 1.0 | standard |
| 7.32882 | 0.216598 | standard |
| 7.6855 | 0.248386 | standard |
| 7.75552 | 0.197115 | standard |
| 8.23776 | 0.261763 | standard |
| 2.32462 | 1.0 | standard |
| 7.32733 | 0.214703 | standard |
| 7.68921 | 0.24279 | standard |
| 7.75294 | 0.19731 | standard |
| 8.23935 | 0.259703 | standard |
| 2.32462 | 1.0 | standard |
| 7.32634 | 0.21365 | standard |
| 7.69229 | 0.239127 | standard |
| 7.75016 | 0.197995 | standard |
| 8.24084 | 0.258838 | standard |
| 2.32462 | 1.0 | standard |
| 7.32356 | 0.210542 | standard |
| 7.70639 | 0.223762 | standard |
| 7.73613 | 0.203004 | standard |
| 8.24719 | 0.254 | standard |
| 2.32462 | 1.0 | standard |
| 7.32308 | 0.209 | standard |
| 7.71128 | 0.218831 | standard |
| 7.73114 | 0.205378 | standard |
| 8.24967 | 0.252888 | standard |
| 2.32462 | 1.0 | standard |
| 7.32278 | 0.209478 | standard |
| 7.71369 | 0.21664 | standard |
| 7.72871 | 0.207145 | standard |
| 8.24937 | 0.252711 | standard |
| 2.32462 | 1.0 | standard |
| 7.32273 | 0.20939 | standard |
| 7.71514 | 0.215555 | standard |
| 7.72717 | 0.207947 | standard |
| 8.24937 | 0.252899 | standard |
| 2.32462 | 1.0 | standard |
| 7.32271 | 0.208895 | standard |
| 7.71619 | 0.213914 | standard |
| 7.72621 | 0.209065 | standard |
| 8.24927 | 0.252899 | standard |
| 2.32462 | 1.0 | standard |
| 7.32264 | 0.208882 | standard |
| 7.71687 | 0.212788 | standard |
| 7.72549 | 0.210316 | standard |
| 8.24932 | 0.252622 | standard |
| 2.32462 | 1.0 | standard |
| 7.32261 | 0.208883 | standard |
| 7.71713 | 0.212311 | standard |
| 7.72515 | 0.210923 | standard |
| 8.24937 | 0.253488 | standard |
| 2.32462 | 1.0 | standard |
| 7.3226 | 0.208539 | standard |
| 7.71738 | 0.211924 | standard |
| 7.7249 | 0.210761 | standard |
| 8.24941 | 0.253296 | standard |
| 2.32462 | 1.0 | standard |
| 7.32255 | 0.209597 | standard |
| 7.71781 | 0.211598 | standard |
| 7.7245 | 0.211597 | standard |
| 8.24932 | 0.254643 | standard |
| 2.32462 | 1.0 | standard |
| 7.32262 | 0.208733 | standard |
| 7.71797 | 0.211482 | standard |
| 7.72433 | 0.211482 | standard |
| 8.24932 | 0.252957 | standard |
| 2.32462 | 1.0 | standard |
| 7.32262 | 0.209049 | standard |
| 7.71814 | 0.211497 | standard |
| 7.72416 | 0.211552 | standard |
| 8.24937 | 0.251482 | standard |
| 2.32462 | 1.0 | standard |
| 7.32262 | 0.209915 | standard |
| 7.7188 | 0.21073 | standard |
| 7.72351 | 0.210743 | standard |
| 8.24932 | 0.25461 | standard |