(1-Methyl-5-phenyl-1H-pyrazol-3-yl)methanol
Simulation outputs:
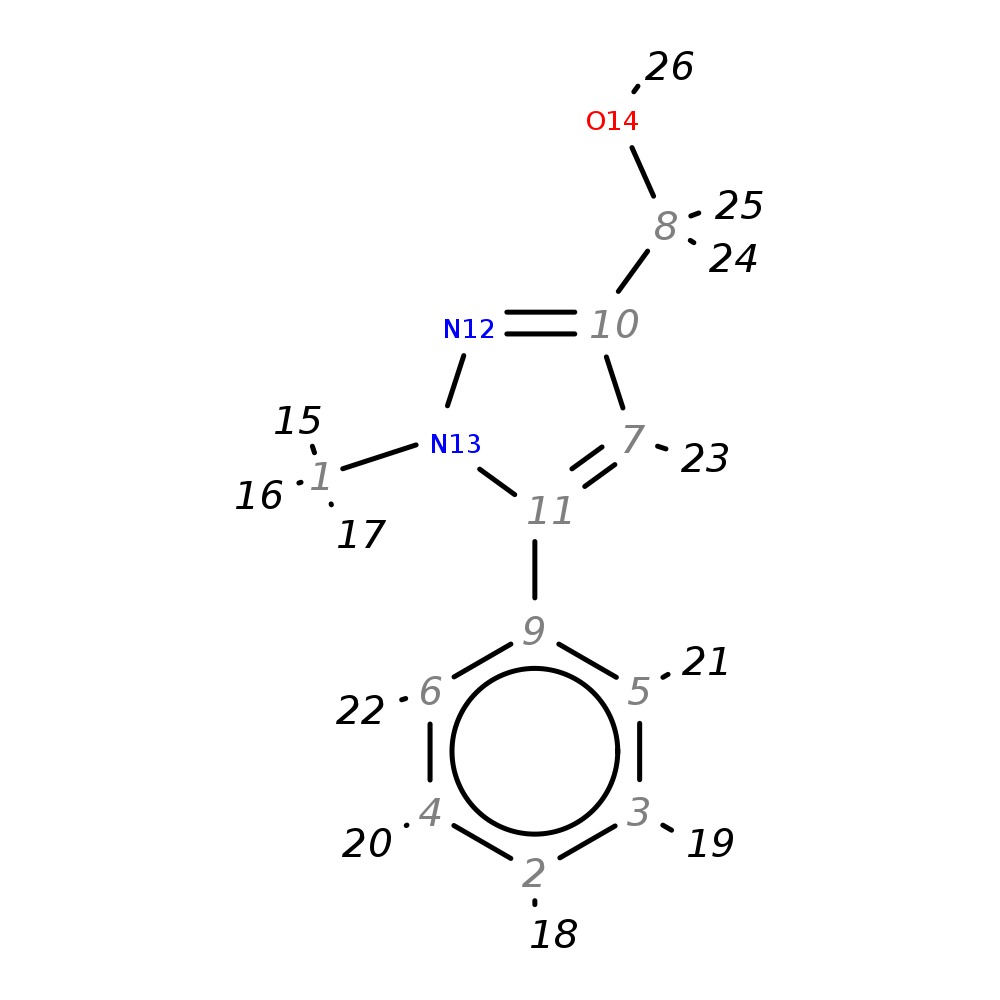
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.00838 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H12N2O/c1-13-11(7-10(8-14)12-13)9-5-3-2-4-6-9/h2-7,14H,8H2,1H3 | |
| Note 1 | 24,25 missing (saturated?) | |
| Note 2 | aromatic coupling - higher field |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| (1-Methyl-5-phenyl-1H-pyrazol-3-yl)methanol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 3.868 | 7.56 | 7.56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 3.878 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 3.878 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 5.686 | 6.81 | 6.81 | 1.5 | 1.5 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 5.936 | 1.5 | 6.55 | 0.5 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 5.936 | 0.5 | 6.55 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 6.395 | 1.5 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.395 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.54 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.712 | 12.55 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.711 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.66522 | standard |
| 6.46506 | 0.337089 | standard |
| 7.50426 | 0.0157323 | standard |
| 7.51065 | 0.0302559 | standard |
| 7.51485 | 0.0437772 | standard |
| 7.51804 | 0.0602538 | standard |
| 7.52562 | 0.0638443 | standard |
| 7.52823 | 0.0669484 | standard |
| 7.53239 | 0.0871287 | standard |
| 7.54279 | 0.118397 | standard |
| 7.55005 | 0.794569 | standard |
| 7.55552 | 0.337963 | standard |
| 7.57336 | 0.01772 | standard |
| 3.83146 | 0.624827 | standard |
| 4.71174 | 0.416952 | standard |
| 6.46506 | 0.209616 | standard |
| 7.54628 | 1.0 | standard |
| 3.83146 | 0.647507 | standard |
| 4.71174 | 0.431815 | standard |
| 6.46506 | 0.217562 | standard |
| 7.54619 | 1.0 | standard |
| 3.83146 | 0.67528 | standard |
| 4.71174 | 0.449431 | standard |
| 6.46506 | 0.227107 | standard |
| 7.54616 | 1.0 | standard |
| 3.83146 | 0.689713 | standard |
| 4.71174 | 0.458862 | standard |
| 6.46506 | 0.232415 | standard |
| 7.54618 | 1.0 | standard |
| 3.83146 | 0.70708 | standard |
| 4.71174 | 0.471318 | standard |
| 6.46506 | 0.238081 | standard |
| 7.54625 | 1.00001 | standard |
| 3.83146 | 0.897408 | standard |
| 4.71174 | 0.598186 | standard |
| 6.46506 | 0.303377 | standard |
| 7.46014 | 0.00681625 | standard |
| 7.54764 | 1.00003 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.666563 | standard |
| 6.46506 | 0.337823 | standard |
| 7.48484 | 0.0126227 | standard |
| 7.50902 | 0.0379183 | standard |
| 7.52588 | 0.0980196 | standard |
| 7.54869 | 0.946592 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.666113 | standard |
| 6.46506 | 0.336959 | standard |
| 7.49835 | 0.0165787 | standard |
| 7.51435 | 0.047466 | standard |
| 7.53707 | 0.171131 | standard |
| 7.54923 | 0.856056 | standard |
| 7.5847 | 0.0130911 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.666287 | standard |
| 6.46506 | 0.33761 | standard |
| 7.50036 | 0.013933 | standard |
| 7.50612 | 0.021244 | standard |
| 7.51298 | 0.0396744 | standard |
| 7.51666 | 0.0551717 | standard |
| 7.52551 | 0.0668833 | standard |
| 7.5338 | 0.121256 | standard |
| 7.5427 | 0.165996 | standard |
| 7.5497 | 0.803636 | standard |
| 7.55583 | 0.342402 | standard |
| 7.57768 | 0.0146367 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.666509 | standard |
| 6.46506 | 0.33735 | standard |
| 7.50426 | 0.0157445 | standard |
| 7.51065 | 0.0302789 | standard |
| 7.51485 | 0.0438112 | standard |
| 7.51804 | 0.0602998 | standard |
| 7.52562 | 0.0638952 | standard |
| 7.52823 | 0.0670014 | standard |
| 7.53239 | 0.0871977 | standard |
| 7.54279 | 0.118492 | standard |
| 7.55005 | 0.795188 | standard |
| 7.55552 | 0.338227 | standard |
| 7.57336 | 0.017734 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.666471 | standard |
| 6.46506 | 0.33619 | standard |
| 7.50687 | 0.0179059 | standard |
| 7.5125 | 0.036318 | standard |
| 7.51611 | 0.0472332 | standard |
| 7.51884 | 0.0638228 | standard |
| 7.52581 | 0.061349 | standard |
| 7.52734 | 0.0629761 | standard |
| 7.53106 | 0.0724008 | standard |
| 7.53844 | 0.0572623 | standard |
| 7.54395 | 0.0929549 | standard |
| 7.55035 | 0.779221 | standard |
| 7.55487 | 0.336594 | standard |
| 7.56788 | 0.0185062 | standard |
| 7.57046 | 0.0216029 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.66548 | standard |
| 6.46506 | 0.3353 | standard |
| 7.5016 | 0.00507813 | standard |
| 7.50795 | 0.0185196 | standard |
| 7.51319 | 0.0370379 | standard |
| 7.51646 | 0.0473783 | standard |
| 7.51917 | 0.0646039 | standard |
| 7.52677 | 0.0626918 | standard |
| 7.53054 | 0.0678488 | standard |
| 7.53783 | 0.045587 | standard |
| 7.55047 | 0.775918 | standard |
| 7.55467 | 0.337861 | standard |
| 7.56922 | 0.023846 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.666788 | standard |
| 6.46506 | 0.336654 | standard |
| 7.50284 | 0.005121 | standard |
| 7.50895 | 0.0192874 | standard |
| 7.51378 | 0.036954 | standard |
| 7.51692 | 0.0488133 | standard |
| 7.51934 | 0.0657076 | standard |
| 7.52156 | 0.0531883 | standard |
| 7.52655 | 0.061583 | standard |
| 7.53004 | 0.0636167 | standard |
| 7.53673 | 0.0354012 | standard |
| 7.54441 | 0.0832661 | standard |
| 7.55058 | 0.776465 | standard |
| 7.55448 | 0.341894 | standard |
| 7.56818 | 0.025133 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.665897 | standard |
| 6.46506 | 0.336095 | standard |
| 7.50484 | 0.00516008 | standard |
| 7.51041 | 0.0202115 | standard |
| 7.51499 | 0.0370616 | standard |
| 7.5174 | 0.0506125 | standard |
| 7.51972 | 0.0667438 | standard |
| 7.52207 | 0.0553703 | standard |
| 7.52354 | 0.0539679 | standard |
| 7.52592 | 0.0622465 | standard |
| 7.52915 | 0.0586284 | standard |
| 7.53519 | 0.0280412 | standard |
| 7.54484 | 0.0759682 | standard |
| 7.55079 | 0.775357 | standard |
| 7.55415 | 0.343406 | standard |
| 7.55605 | 0.258202 | standard |
| 7.56654 | 0.0272544 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.666287 | standard |
| 6.46506 | 0.336193 | standard |
| 7.50571 | 0.00517303 | standard |
| 7.51099 | 0.0204806 | standard |
| 7.51585 | 0.0373592 | standard |
| 7.51762 | 0.0513101 | standard |
| 7.51986 | 0.0666843 | standard |
| 7.52218 | 0.0566125 | standard |
| 7.52324 | 0.0539955 | standard |
| 7.52571 | 0.0620118 | standard |
| 7.52884 | 0.0570992 | standard |
| 7.53456 | 0.0259985 | standard |
| 7.54523 | 0.0740405 | standard |
| 7.55088 | 0.77275 | standard |
| 7.55405 | 0.344563 | standard |
| 7.55604 | 0.258087 | standard |
| 7.56586 | 0.0284001 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.665731 | standard |
| 6.46506 | 0.33571 | standard |
| 7.50646 | 0.00516529 | standard |
| 7.51159 | 0.0211238 | standard |
| 7.51505 | 0.0357402 | standard |
| 7.51788 | 0.0519954 | standard |
| 7.51994 | 0.067145 | standard |
| 7.52223 | 0.0573768 | standard |
| 7.52555 | 0.0628232 | standard |
| 7.52843 | 0.0556551 | standard |
| 7.53396 | 0.0239399 | standard |
| 7.54551 | 0.073952 | standard |
| 7.55095 | 0.768455 | standard |
| 7.5539 | 0.344248 | standard |
| 7.55601 | 0.259567 | standard |
| 7.56525 | 0.0294066 | standard |
| 3.83146 | 1.0 | standard |
| 4.71174 | 0.665352 | standard |
| 6.46506 | 0.335381 | standard |
| 7.51112 | 0.00700538 | standard |
| 7.51397 | 0.0228639 | standard |
| 7.51599 | 0.0374445 | standard |
| 7.5186 | 0.0541937 | standard |
| 7.52044 | 0.0701518 | standard |
| 7.52235 | 0.0617827 | standard |
| 7.52484 | 0.0643413 | standard |
| 7.52692 | 0.0497437 | standard |
| 7.52897 | 0.0203192 | standard |
| 7.53141 | 0.0176084 | standard |
| 7.54681 | 0.0711313 | standard |
| 7.55131 | 0.764224 | standard |
| 7.55335 | 0.352577 | standard |
| 7.5556 | 0.261608 | standard |
| 7.5625 | 0.0347519 | standard |