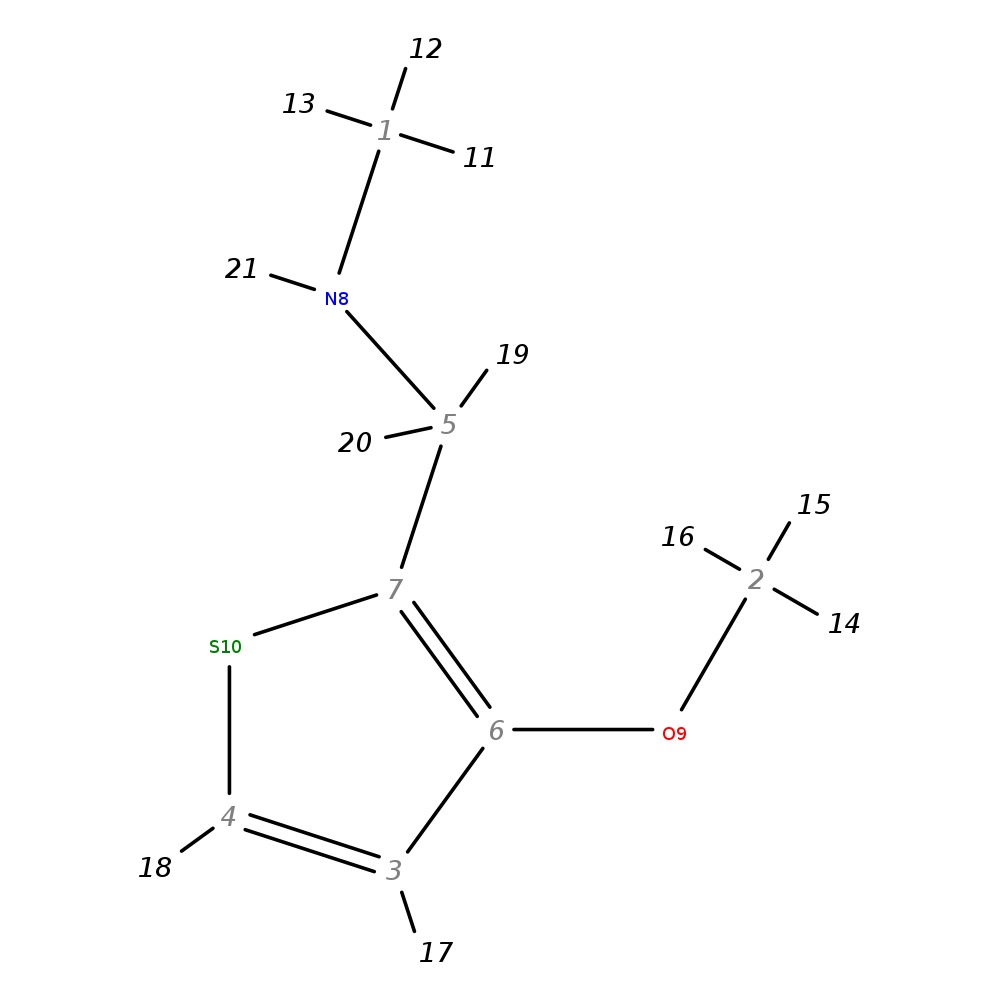
N-methyl-(3-methoxythien-2-yl)methylamine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01582 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H11NOS/c1-8-5-7-6(9-2)3-4-10-7/h3-4,8H,5H2,1-2H3 | |
| Note 1 | 17?18 | |
| Note 2 | 11,12,13?14,15,16 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-methyl-(3-methoxythien-2-yl)methylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 2.686 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 2.686 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 2.686 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 3.908 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 3.908 | -14.0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 3.908 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 7.065 | 5.862 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.49 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.295 | -14.0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.295 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.68576 | 0.999684 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.665864 | standard |
| 7.06048 | 0.170507 | standard |
| 7.07018 | 0.17237 | standard |
| 7.485 | 0.171238 | standard |
| 7.4948 | 0.170385 | standard |
| 2.68576 | 0.997202 | standard |
| 3.90764 | 1.0 | standard |
| 4.29515 | 0.667497 | standard |
| 6.98008 | 0.119039 | standard |
| 7.12628 | 0.225426 | standard |
| 7.42889 | 0.225407 | standard |
| 7.5752 | 0.119002 | standard |
| 2.68576 | 0.997764 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.66688 | standard |
| 7.01111 | 0.135422 | standard |
| 7.10863 | 0.208289 | standard |
| 7.44656 | 0.20824 | standard |
| 7.54408 | 0.134014 | standard |
| 2.68576 | 0.998618 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.666058 | standard |
| 7.02574 | 0.143681 | standard |
| 7.09882 | 0.198875 | standard |
| 7.45637 | 0.199102 | standard |
| 7.52954 | 0.143401 | standard |
| 2.68576 | 0.999285 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.666305 | standard |
| 7.03039 | 0.146564 | standard |
| 7.09545 | 0.196094 | standard |
| 7.45983 | 0.196108 | standard |
| 7.52479 | 0.146971 | standard |
| 2.68576 | 0.999053 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.666964 | standard |
| 7.03415 | 0.149326 | standard |
| 7.09267 | 0.193666 | standard |
| 7.46261 | 0.193662 | standard |
| 7.52113 | 0.149321 | standard |
| 2.68576 | 0.999666 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.666599 | standard |
| 7.05028 | 0.160179 | standard |
| 7.0795 | 0.182579 | standard |
| 7.47569 | 0.18255 | standard |
| 7.505 | 0.160238 | standard |
| 2.68576 | 0.998979 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.666046 | standard |
| 7.05543 | 0.164536 | standard |
| 7.07494 | 0.178212 | standard |
| 7.48034 | 0.178164 | standard |
| 7.49985 | 0.163189 | standard |
| 2.68576 | 0.997968 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.665365 | standard |
| 7.05791 | 0.167609 | standard |
| 7.07256 | 0.173475 | standard |
| 7.48262 | 0.17506 | standard |
| 7.49728 | 0.16608 | standard |
| 2.68576 | 0.999883 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.665803 | standard |
| 7.05949 | 0.16947 | standard |
| 7.07118 | 0.172696 | standard |
| 7.48411 | 0.173263 | standard |
| 7.49579 | 0.169299 | standard |
| 2.68576 | 0.999061 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.665339 | standard |
| 7.06048 | 0.170471 | standard |
| 7.07018 | 0.171292 | standard |
| 7.485 | 0.172179 | standard |
| 7.4948 | 0.169378 | standard |
| 2.68576 | 1.0 | standard |
| 3.90764 | 0.998671 | standard |
| 4.29516 | 0.665357 | standard |
| 7.06117 | 0.171477 | standard |
| 7.06949 | 0.171442 | standard |
| 7.48569 | 0.171334 | standard |
| 7.49411 | 0.171369 | standard |
| 2.68576 | 0.999624 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.66575 | standard |
| 7.06147 | 0.171313 | standard |
| 7.06929 | 0.170823 | standard |
| 7.48599 | 0.171212 | standard |
| 7.49381 | 0.170854 | standard |
| 2.68576 | 0.999307 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.667131 | standard |
| 7.06167 | 0.171368 | standard |
| 7.069 | 0.171285 | standard |
| 7.48619 | 0.171367 | standard |
| 7.49351 | 0.171302 | standard |
| 2.68576 | 1.0 | standard |
| 3.90764 | 0.999944 | standard |
| 4.29516 | 0.666508 | standard |
| 7.06206 | 0.171317 | standard |
| 7.0686 | 0.171317 | standard |
| 7.48658 | 0.171368 | standard |
| 7.49312 | 0.171316 | standard |
| 2.68576 | 1.0 | standard |
| 3.90764 | 0.999483 | standard |
| 4.29516 | 0.664907 | standard |
| 7.06226 | 0.171313 | standard |
| 7.0684 | 0.170402 | standard |
| 7.48678 | 0.171354 | standard |
| 7.49292 | 0.171382 | standard |
| 2.68576 | 0.998664 | standard |
| 3.90764 | 1.0 | standard |
| 4.29516 | 0.665419 | standard |
| 7.06246 | 0.171281 | standard |
| 7.0683 | 0.169495 | standard |
| 7.48698 | 0.171385 | standard |
| 7.49282 | 0.170846 | standard |
| 2.68576 | 1.0 | standard |
| 3.90764 | 0.999858 | standard |
| 4.29516 | 0.666383 | standard |
| 7.06315 | 0.171454 | standard |
| 7.06761 | 0.171455 | standard |
| 7.48757 | 0.171252 | standard |
| 7.49213 | 0.171252 | standard |