N-methyl-4-(1H-pyrazol-1-ylmethyl)benzylamine
Simulation outputs:
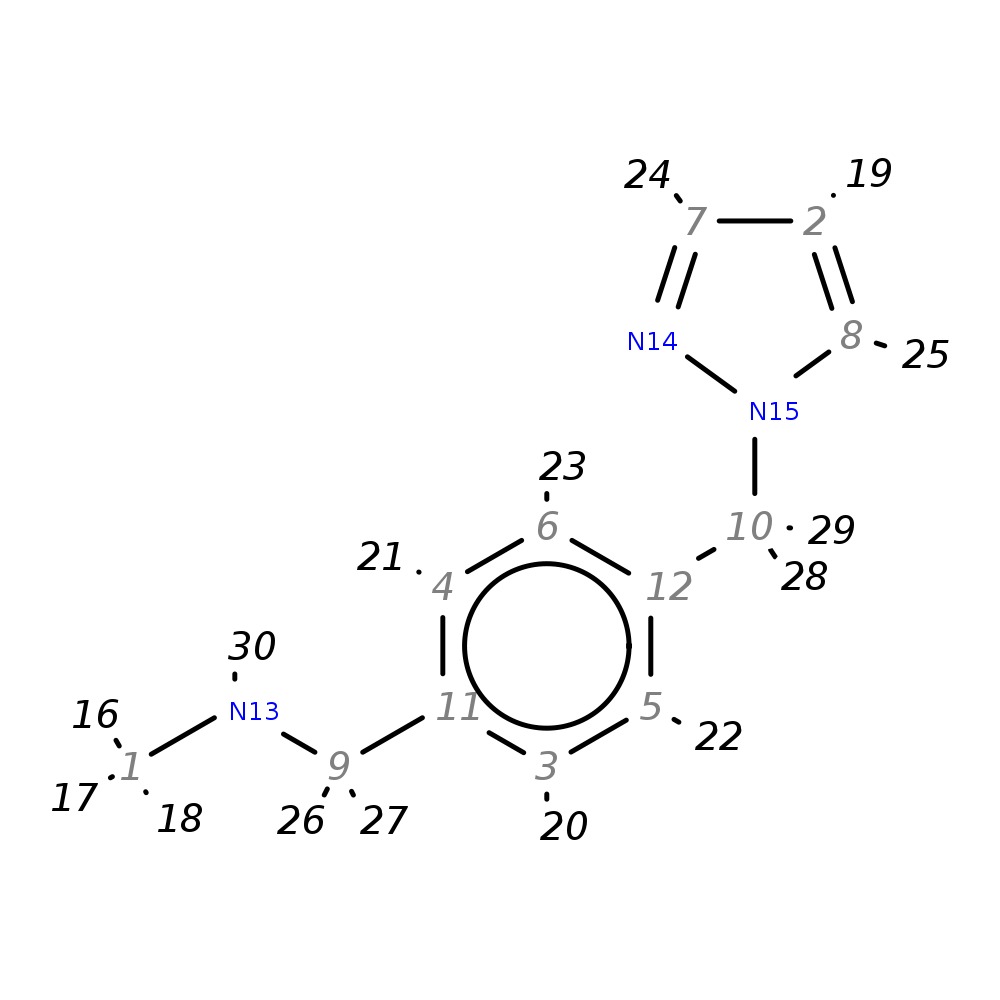
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01543 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H15N3/c1-13-9-11-3-5-12(6-4-11)10-15-8-2-7-14-15/h2-8,13H,9-10H2,1H3 | |
| Note 1 | 24?25 | |
| Note 2 | 20,21?22,23 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-methyl-4-(1H-pyrazol-1-ylmethyl)benzylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 2.699 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 2.699 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 2.699 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 6.41 | 0 | 0 | 0 | 0 | 2.211 | 2.014 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 7.317 | 1.5 | 7.765 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 7.317 | 0 | 7.765 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 7.447 | 1.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.447 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.594 | 0 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.803 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.199 | -14.0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.199 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.397 | -14.0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.397 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666501 | standard |
| 5.39672 | 0.666547 | standard |
| 6.41022 | 0.160026 | standard |
| 7.31006 | 0.206535 | standard |
| 7.32271 | 0.247433 | standard |
| 7.44126 | 0.247472 | standard |
| 7.45391 | 0.206587 | standard |
| 7.59254 | 0.161214 | standard |
| 7.59555 | 0.161209 | standard |
| 7.80131 | 0.166326 | standard |
| 7.80386 | 0.166325 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.66694 | standard |
| 5.39672 | 0.667868 | standard |
| 6.40951 | 0.162354 | standard |
| 7.17072 | 0.0612071 | standard |
| 7.38223 | 0.725786 | standard |
| 7.57222 | 0.225443 | standard |
| 7.6116 | 0.216323 | standard |
| 7.78273 | 0.184935 | standard |
| 7.82079 | 0.176629 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.667164 | standard |
| 5.39672 | 0.667088 | standard |
| 6.4098 | 0.161249 | standard |
| 7.22542 | 0.083603 | standard |
| 7.35886 | 0.469023 | standard |
| 7.40516 | 0.470418 | standard |
| 7.57684 | 0.196343 | standard |
| 7.60859 | 0.177777 | standard |
| 7.78994 | 0.173627 | standard |
| 7.81533 | 0.171805 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666267 | standard |
| 5.39672 | 0.666329 | standard |
| 6.40999 | 0.160849 | standard |
| 7.25218 | 0.104291 | standard |
| 7.35056 | 0.38857 | standard |
| 7.41342 | 0.389643 | standard |
| 7.51243 | 0.114925 | standard |
| 7.5825 | 0.174385 | standard |
| 7.60539 | 0.170365 | standard |
| 7.79311 | 0.170114 | standard |
| 7.81216 | 0.16965 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666627 | standard |
| 5.39672 | 0.666461 | standard |
| 6.41003 | 0.160775 | standard |
| 7.26079 | 0.11335 | standard |
| 7.34777 | 0.367467 | standard |
| 7.41627 | 0.36812 | standard |
| 7.50344 | 0.120067 | standard |
| 7.58386 | 0.170615 | standard |
| 7.60426 | 0.168232 | standard |
| 7.79417 | 0.169296 | standard |
| 7.81114 | 0.168971 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.665974 | standard |
| 5.39672 | 0.666386 | standard |
| 6.41001 | 0.160595 | standard |
| 7.26739 | 0.12139 | standard |
| 7.34536 | 0.35152 | standard |
| 7.4186 | 0.352317 | standard |
| 7.49669 | 0.126021 | standard |
| 7.58492 | 0.168247 | standard |
| 7.60323 | 0.166943 | standard |
| 7.79502 | 0.168815 | standard |
| 7.81022 | 0.16858 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666615 | standard |
| 5.39672 | 0.666832 | standard |
| 6.41016 | 0.160926 | standard |
| 7.29478 | 0.167436 | standard |
| 7.33317 | 0.287199 | standard |
| 7.43087 | 0.287583 | standard |
| 7.46919 | 0.168378 | standard |
| 7.58951 | 0.163115 | standard |
| 7.59857 | 0.162984 | standard |
| 7.79888 | 0.167886 | standard |
| 7.80639 | 0.167857 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666142 | standard |
| 5.39672 | 0.666069 | standard |
| 6.41015 | 0.160365 | standard |
| 7.30273 | 0.186351 | standard |
| 7.32822 | 0.267062 | standard |
| 7.43575 | 0.26741 | standard |
| 7.46124 | 0.186596 | standard |
| 7.59095 | 0.161074 | standard |
| 7.59714 | 0.161036 | standard |
| 7.80003 | 0.1666 | standard |
| 7.80514 | 0.166591 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666275 | standard |
| 5.39672 | 0.665976 | standard |
| 6.41015 | 0.160582 | standard |
| 7.30646 | 0.195959 | standard |
| 7.32554 | 0.256902 | standard |
| 7.43843 | 0.256985 | standard |
| 7.45751 | 0.196085 | standard |
| 7.59182 | 0.161977 | standard |
| 7.59627 | 0.161962 | standard |
| 7.80072 | 0.16641 | standard |
| 7.80455 | 0.166406 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.66694 | standard |
| 5.39672 | 0.666763 | standard |
| 6.41022 | 0.160517 | standard |
| 7.30859 | 0.201883 | standard |
| 7.32388 | 0.250844 | standard |
| 7.44008 | 0.250899 | standard |
| 7.45538 | 0.20196 | standard |
| 7.59219 | 0.160583 | standard |
| 7.5959 | 0.160575 | standard |
| 7.80108 | 0.166102 | standard |
| 7.80418 | 0.1661 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666751 | standard |
| 5.39672 | 0.666507 | standard |
| 6.41022 | 0.160041 | standard |
| 7.31006 | 0.206554 | standard |
| 7.32271 | 0.247456 | standard |
| 7.44126 | 0.247496 | standard |
| 7.45391 | 0.206606 | standard |
| 7.59254 | 0.161229 | standard |
| 7.59555 | 0.161225 | standard |
| 7.80131 | 0.166342 | standard |
| 7.80386 | 0.166341 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666946 | standard |
| 5.39672 | 0.666824 | standard |
| 6.41016 | 0.160161 | standard |
| 7.31103 | 0.208629 | standard |
| 7.32193 | 0.24369 | standard |
| 7.44204 | 0.243683 | standard |
| 7.45294 | 0.20956 | standard |
| 7.59281 | 0.162551 | standard |
| 7.59529 | 0.162548 | standard |
| 7.80155 | 0.166666 | standard |
| 7.80372 | 0.166666 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.66681 | standard |
| 5.39672 | 0.666481 | standard |
| 6.41022 | 0.160636 | standard |
| 7.31142 | 0.210268 | standard |
| 7.32154 | 0.243584 | standard |
| 7.44243 | 0.24361 | standard |
| 7.45255 | 0.210301 | standard |
| 7.59288 | 0.162537 | standard |
| 7.59521 | 0.162535 | standard |
| 7.80163 | 0.166982 | standard |
| 7.80364 | 0.166981 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666293 | standard |
| 5.39672 | 0.666469 | standard |
| 6.41021 | 0.159983 | standard |
| 7.31181 | 0.211164 | standard |
| 7.32124 | 0.242351 | standard |
| 7.44273 | 0.242273 | standard |
| 7.45227 | 0.210037 | standard |
| 7.59297 | 0.162887 | standard |
| 7.59512 | 0.162885 | standard |
| 7.80172 | 0.167754 | standard |
| 7.80355 | 0.167754 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666056 | standard |
| 5.39672 | 0.666346 | standard |
| 6.41021 | 0.159645 | standard |
| 7.31229 | 0.212054 | standard |
| 7.32075 | 0.241601 | standard |
| 7.44322 | 0.241613 | standard |
| 7.45168 | 0.211669 | standard |
| 7.59302 | 0.160249 | standard |
| 7.59507 | 0.160248 | standard |
| 7.80179 | 0.168194 | standard |
| 7.80338 | 0.168193 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.66657 | standard |
| 5.39672 | 0.666485 | standard |
| 6.41022 | 0.160182 | standard |
| 7.31258 | 0.212954 | standard |
| 7.32056 | 0.239683 | standard |
| 7.44341 | 0.2397 | standard |
| 7.45139 | 0.212974 | standard |
| 7.59312 | 0.161998 | standard |
| 7.59497 | 0.161997 | standard |
| 7.80187 | 0.167718 | standard |
| 7.8034 | 0.167718 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.666138 | standard |
| 5.39672 | 0.666202 | standard |
| 6.41021 | 0.160942 | standard |
| 7.31278 | 0.214131 | standard |
| 7.32037 | 0.239602 | standard |
| 7.4436 | 0.239617 | standard |
| 7.45119 | 0.214149 | standard |
| 7.59308 | 0.158982 | standard |
| 7.59501 | 0.158981 | standard |
| 7.80185 | 0.167489 | standard |
| 7.80332 | 0.167489 | standard |
| 2.69895 | 1.0 | standard |
| 4.19934 | 0.66742 | standard |
| 5.39672 | 0.667084 | standard |
| 6.41021 | 0.161108 | standard |
| 7.31372 | 0.21673 | standard |
| 7.31959 | 0.234828 | standard |
| 7.44437 | 0.235446 | standard |
| 7.45032 | 0.215866 | standard |
| 7.59336 | 0.161357 | standard |
| 7.59474 | 0.161357 | standard |
| 7.80197 | 0.165059 | standard |
| 7.8032 | 0.165058 | standard |