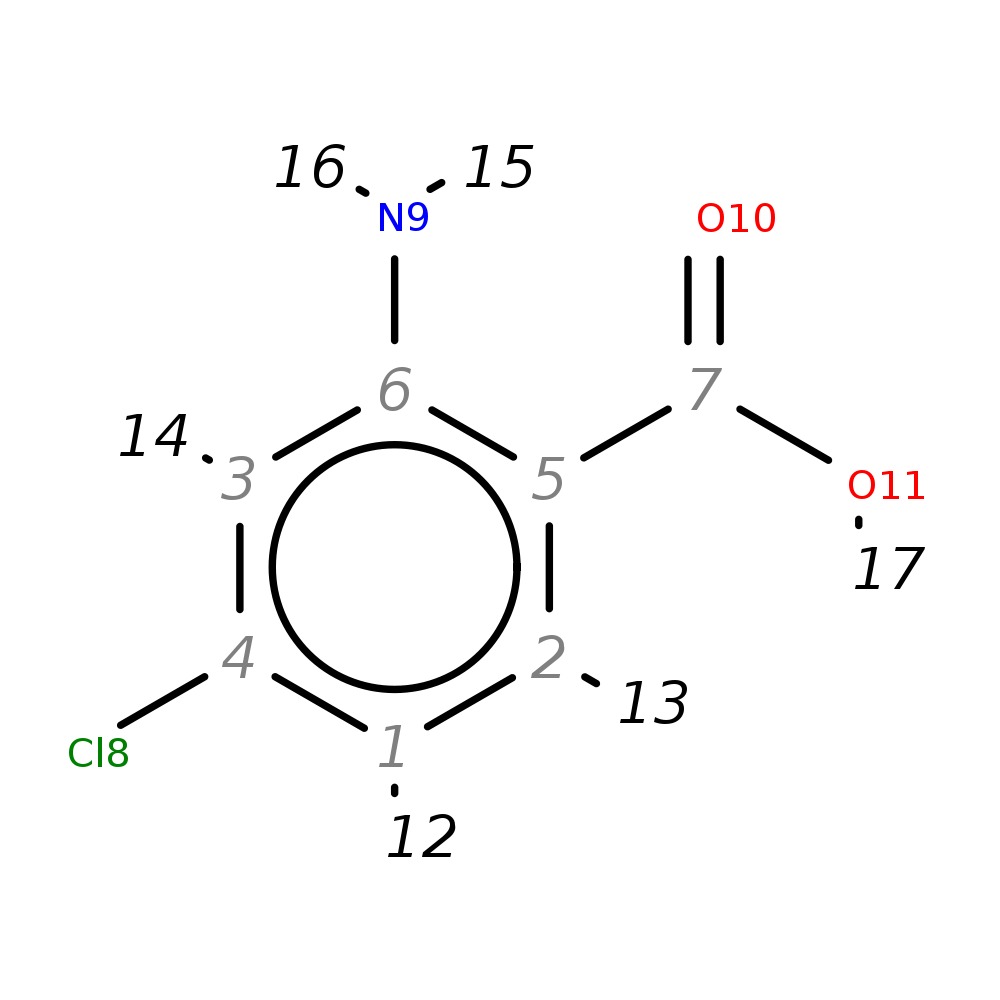
2-Amino-4-chlorobenzoic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03385 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H6ClNO2/c8-4-1-2-5(7(10)11)6(9)3-4/h1-3H,9H2,(H,10,11) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-Amino-4-chlorobenzoic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | |
|---|---|---|---|
| 12 | 6.757 | 8.921 | 2.03 |
| 13 | 0 | 7.645 | 0 |
| 14 | 0 | 0 | 6.862 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 6.7479 | 0.488984 | standard |
| 6.75098 | 0.512758 | standard |
| 6.76265 | 0.488317 | standard |
| 6.76586 | 0.505939 | standard |
| 6.86025 | 1.00036 | standard |
| 6.86333 | 0.95905 | standard |
| 7.63744 | 0.824092 | standard |
| 7.65231 | 0.824091 | standard |
| 6.60843 | 0.152447 | standard |
| 6.65293 | 0.208141 | standard |
| 6.85712 | 1.0 | standard |
| 7.55078 | 0.352775 | standard |
| 7.76541 | 0.221473 | standard |
| 6.66058 | 0.214576 | standard |
| 6.69089 | 0.276893 | standard |
| 6.84557 | 1.0 | standard |
| 7.57906 | 0.511835 | standard |
| 7.72287 | 0.369719 | standard |
| 6.68575 | 0.315514 | standard |
| 6.70868 | 0.391445 | standard |
| 6.81964 | 0.683357 | standard |
| 6.85137 | 1.0 | standard |
| 6.87291 | 0.661076 | standard |
| 7.59351 | 0.716192 | standard |
| 7.70309 | 0.559769 | standard |
| 6.69405 | 0.340837 | standard |
| 6.71438 | 0.41608 | standard |
| 6.79358 | 0.40119 | standard |
| 6.81279 | 0.637273 | standard |
| 6.85246 | 1.0 | standard |
| 6.87212 | 0.707166 | standard |
| 7.59867 | 0.752945 | standard |
| 7.69644 | 0.604458 | standard |
| 6.7005 | 0.358186 | standard |
| 6.71889 | 0.431627 | standard |
| 6.78956 | 0.413227 | standard |
| 6.80761 | 0.610043 | standard |
| 6.85329 | 1.0 | standard |
| 6.87131 | 0.74472 | standard |
| 7.60293 | 0.774193 | standard |
| 7.69118 | 0.640463 | standard |
| 6.72934 | 0.431108 | standard |
| 6.73857 | 0.482562 | standard |
| 6.77377 | 0.468575 | standard |
| 6.78311 | 0.541476 | standard |
| 6.85724 | 1.0 | standard |
| 6.86651 | 0.876712 | standard |
| 7.62311 | 0.825161 | standard |
| 7.66763 | 0.74868 | standard |
| 6.73866 | 0.455042 | standard |
| 6.74482 | 0.494038 | standard |
| 6.76829 | 0.483237 | standard |
| 6.77457 | 0.526785 | standard |
| 6.85873 | 1.0 | standard |
| 6.86488 | 0.918208 | standard |
| 7.6302 | 0.831904 | standard |
| 7.6599 | 0.779286 | standard |
| 6.74321 | 0.466035 | standard |
| 6.74796 | 0.498218 | standard |
| 6.76553 | 0.494092 | standard |
| 6.77015 | 0.524237 | standard |
| 6.85941 | 1.00001 | standard |
| 6.86409 | 0.937652 | standard |
| 7.63377 | 0.836805 | standard |
| 7.65608 | 0.796858 | standard |
| 6.74605 | 0.475485 | standard |
| 6.74977 | 0.502527 | standard |
| 6.76389 | 0.497163 | standard |
| 6.76761 | 0.519862 | standard |
| 6.85988 | 1.00029 | standard |
| 6.8636 | 0.950698 | standard |
| 7.63595 | 0.837619 | standard |
| 7.6538 | 0.805454 | standard |
| 6.7479 | 0.488985 | standard |
| 6.75098 | 0.512759 | standard |
| 6.76265 | 0.488317 | standard |
| 6.76586 | 0.505939 | standard |
| 6.86025 | 1.00036 | standard |
| 6.86333 | 0.95905 | standard |
| 7.63744 | 0.824088 | standard |
| 7.65231 | 0.824088 | standard |
| 6.74917 | 0.491657 | standard |
| 6.75179 | 0.512744 | standard |
| 6.76184 | 0.490534 | standard |
| 6.76459 | 0.504813 | standard |
| 6.86043 | 1.00015 | standard |
| 6.86306 | 0.964873 | standard |
| 7.63843 | 0.825285 | standard |
| 7.65122 | 0.825284 | standard |
| 6.74964 | 0.487483 | standard |
| 6.7522 | 0.50806 | standard |
| 6.76155 | 0.496633 | standard |
| 6.76399 | 0.509393 | standard |
| 6.86053 | 1.00008 | standard |
| 6.86296 | 0.967201 | standard |
| 7.63893 | 0.825743 | standard |
| 7.65083 | 0.825743 | standard |
| 6.75013 | 0.488116 | standard |
| 6.7525 | 0.507596 | standard |
| 6.76123 | 0.492185 | standard |
| 6.7636 | 0.503831 | standard |
| 6.86062 | 1.00005 | standard |
| 6.86287 | 0.969536 | standard |
| 7.63923 | 0.823642 | standard |
| 7.65043 | 0.823643 | standard |
| 6.75093 | 0.501746 | standard |
| 6.7529 | 0.519089 | standard |
| 6.76084 | 0.505598 | standard |
| 6.76281 | 0.515469 | standard |
| 6.8607 | 1.00004 | standard |
| 6.86279 | 0.972103 | standard |
| 7.63992 | 0.83334 | standard |
| 7.64974 | 0.833339 | standard |
| 6.7512 | 0.498606 | standard |
| 6.75321 | 0.503335 | standard |
| 6.76063 | 0.50321 | standard |
| 6.76252 | 0.512233 | standard |
| 6.86081 | 1.0 | standard |
| 6.86269 | 0.986502 | standard |
| 7.64012 | 0.830178 | standard |
| 7.64954 | 0.830178 | standard |
| 6.7515 | 0.512608 | standard |
| 6.7533 | 0.517088 | standard |
| 6.76043 | 0.510706 | standard |
| 6.76223 | 0.519218 | standard |
| 6.86079 | 1.00024 | standard |
| 6.86271 | 0.98688 | standard |
| 7.64037 | 0.840923 | standard |
| 7.64924 | 0.84301 | standard |
| 6.75277 | 0.499515 | standard |
| 6.75422 | 0.504283 | standard |
| 6.7596 | 0.504289 | standard |
| 6.76106 | 0.499526 | standard |
| 6.86109 | 1.00046 | standard |
| 6.86242 | 1.00046 | standard |
| 7.64141 | 0.830874 | standard |
| 7.64825 | 0.830874 | standard |