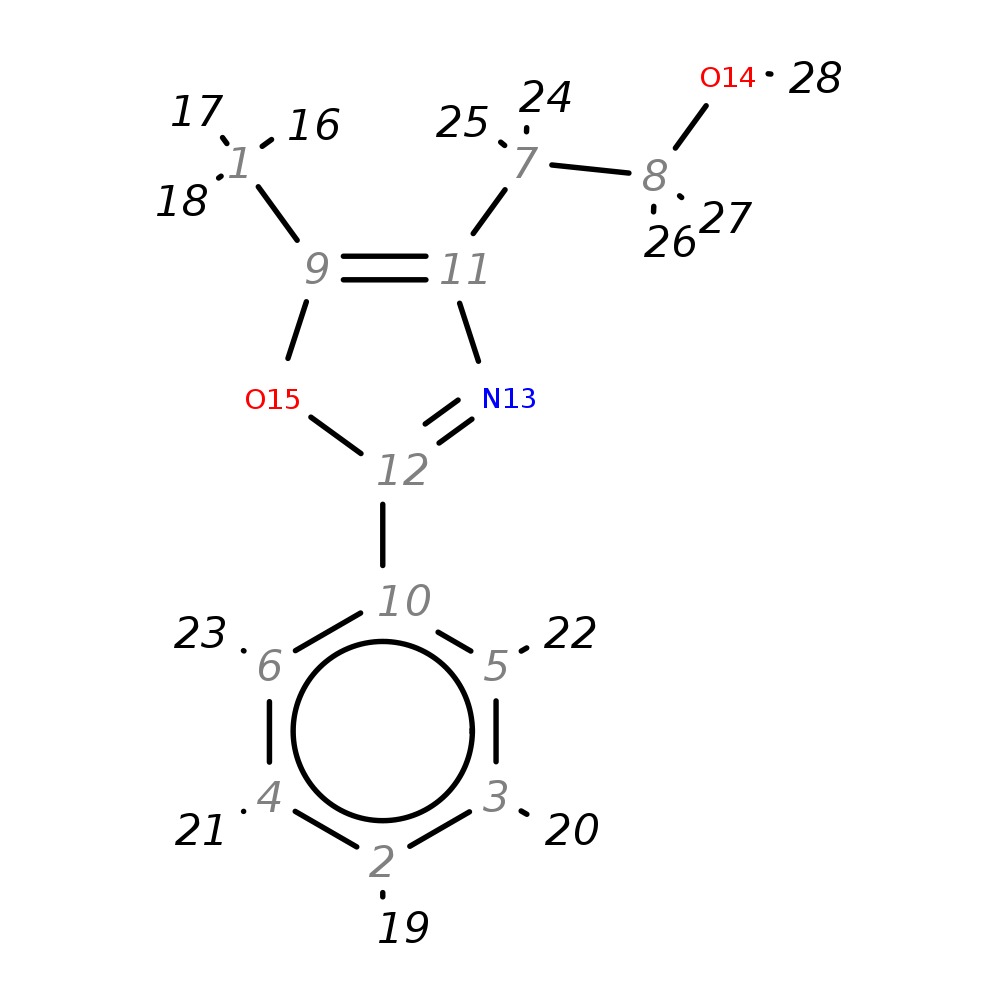
2-(5-Methyl-2-phenyl-1,3-oxazol-4-yl)ethan-1-ol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.04691 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H13NO2/c1-9-11(7-8-14)13-12(15-9)10-5-3-2-4-6-10/h2-6,14H,7-8H2,1H3 | |
| Note 1 | 24,25?26,27 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(5-Methyl-2-phenyl-1,3-oxazol-4-yl)ethan-1-ol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 2.322 | 6.67 | 6.67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 2.324 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 2.324 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 5.657 | 6.81 | 6.81 | 1.5 | 1.5 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 6.633 | 1.5 | 6.55 | 0.5 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 6.633 | 0.5 | 6.55 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 7.482 | 1.5 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.482 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.908 | 15.39 | 6.82 | 6.82 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.912 | 6.82 | 6.82 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.954 | 12.21 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.973 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.35567 | 1.0 | standard |
| 2.73184 | 0.174729 | standard |
| 2.74304 | 0.340437 | standard |
| 2.75424 | 0.174727 | standard |
| 3.82974 | 0.174695 | standard |
| 3.84093 | 0.339861 | standard |
| 3.85218 | 0.174194 | standard |
| 7.54331 | 0.295513 | standard |
| 7.54801 | 0.274809 | standard |
| 7.55471 | 0.24917 | standard |
| 7.5602 | 0.0865871 | standard |
| 7.57132 | 0.0127624 | standard |
| 7.9599 | 0.134414 | standard |
| 7.96324 | 0.147683 | standard |
| 7.97007 | 0.17113 | standard |
| 7.97752 | 0.130775 | standard |
| 2.35568 | 1.0 | standard |
| 2.56798 | 0.115165 | standard |
| 2.7356 | 0.270005 | standard |
| 2.89688 | 0.200411 | standard |
| 3.6871 | 0.198395 | standard |
| 3.84832 | 0.265898 | standard |
| 4.01367 | 0.101971 | standard |
| 7.43674 | 0.162008 | standard |
| 7.53404 | 0.240643 | standard |
| 7.61949 | 0.371575 | standard |
| 7.77526 | 0.117779 | standard |
| 7.81116 | 0.125534 | standard |
| 7.86476 | 0.21495 | standard |
| 7.99674 | 0.122866 | standard |
| 8.13897 | 0.0660512 | standard |
| 2.35567 | 1.0 | standard |
| 2.62607 | 0.130992 | standard |
| 2.73795 | 0.301498 | standard |
| 2.84901 | 0.195223 | standard |
| 3.735 | 0.193917 | standard |
| 3.84604 | 0.298905 | standard |
| 3.95787 | 0.12754 | standard |
| 7.4806 | 0.198768 | standard |
| 7.54108 | 0.257557 | standard |
| 7.59924 | 0.332778 | standard |
| 7.89331 | 0.188724 | standard |
| 7.98305 | 0.133881 | standard |
| 8.03711 | 0.0949352 | standard |
| 8.07302 | 0.0843184 | standard |
| 2.35567 | 1.0 | standard |
| 2.65605 | 0.142756 | standard |
| 2.73999 | 0.317861 | standard |
| 2.82381 | 0.192295 | standard |
| 3.76027 | 0.191678 | standard |
| 3.84399 | 0.316417 | standard |
| 3.92792 | 0.141103 | standard |
| 7.49999 | 0.218881 | standard |
| 7.54408 | 0.265337 | standard |
| 7.58799 | 0.315545 | standard |
| 7.90962 | 0.174327 | standard |
| 7.93321 | 0.143312 | standard |
| 7.97829 | 0.138288 | standard |
| 8.01841 | 0.101457 | standard |
| 8.04354 | 0.0963304 | standard |
| 2.35567 | 1.0 | standard |
| 2.66602 | 0.146506 | standard |
| 2.74062 | 0.322263 | standard |
| 2.81518 | 0.191108 | standard |
| 3.76884 | 0.190263 | standard |
| 3.84336 | 0.32124 | standard |
| 3.918 | 0.145549 | standard |
| 7.50608 | 0.225789 | standard |
| 7.54494 | 0.26784 | standard |
| 7.5841 | 0.309043 | standard |
| 7.91543 | 0.16861 | standard |
| 7.9369 | 0.141901 | standard |
| 7.97694 | 0.139983 | standard |
| 8.01221 | 0.1037 | standard |
| 8.03425 | 0.0999026 | standard |
| 2.35567 | 1.0 | standard |
| 2.67387 | 0.149714 | standard |
| 2.74109 | 0.325526 | standard |
| 2.80824 | 0.189716 | standard |
| 3.77578 | 0.189609 | standard |
| 3.84294 | 0.324072 | standard |
| 3.9101 | 0.148781 | standard |
| 7.51081 | 0.230982 | standard |
| 7.53047 | 0.205513 | standard |
| 7.54565 | 0.269277 | standard |
| 7.58086 | 0.304662 | standard |
| 7.92024 | 0.164769 | standard |
| 7.93941 | 0.141038 | standard |
| 7.97584 | 0.141032 | standard |
| 8.00743 | 0.105258 | standard |
| 8.02703 | 0.103064 | standard |
| 2.35567 | 1.0 | standard |
| 2.70896 | 0.165188 | standard |
| 2.74257 | 0.336357 | standard |
| 2.77623 | 0.181012 | standard |
| 3.80779 | 0.181246 | standard |
| 3.8414 | 0.3357 | standard |
| 3.87506 | 0.165289 | standard |
| 7.53092 | 0.25618 | standard |
| 7.53921 | 0.222005 | standard |
| 7.54771 | 0.272284 | standard |
| 7.5657 | 0.280732 | standard |
| 7.94323 | 0.146531 | standard |
| 7.95323 | 0.137198 | standard |
| 7.97214 | 0.147062 | standard |
| 7.98731 | 0.111031 | standard |
| 7.99629 | 0.118303 | standard |
| 2.35567 | 1.0 | standard |
| 2.72045 | 0.17093 | standard |
| 2.74287 | 0.33862 | standard |
| 2.76533 | 0.177063 | standard |
| 3.81869 | 0.177364 | standard |
| 3.84113 | 0.338695 | standard |
| 3.86357 | 0.171169 | standard |
| 7.53726 | 0.268164 | standard |
| 7.54274 | 0.228586 | standard |
| 7.54802 | 0.271456 | standard |
| 7.56035 | 0.270946 | standard |
| 7.59271 | 0.010757 | standard |
| 7.95141 | 0.139988 | standard |
| 7.95825 | 0.138823 | standard |
| 7.97145 | 0.152796 | standard |
| 7.98089 | 0.111477 | standard |
| 7.98677 | 0.124155 | standard |
| 2.35567 | 1.0 | standard |
| 2.72615 | 0.173919 | standard |
| 2.74299 | 0.338979 | standard |
| 2.75978 | 0.174894 | standard |
| 3.82419 | 0.174823 | standard |
| 3.84103 | 0.33859 | standard |
| 3.85788 | 0.173556 | standard |
| 7.54028 | 0.277144 | standard |
| 7.54804 | 0.270108 | standard |
| 7.55754 | 0.265508 | standard |
| 7.58214 | 0.0113145 | standard |
| 7.9556 | 0.136657 | standard |
| 7.96081 | 0.141048 | standard |
| 7.97092 | 0.159494 | standard |
| 7.97797 | 0.111855 | standard |
| 7.98203 | 0.127438 | standard |
| 2.35567 | 1.0 | standard |
| 2.72956 | 0.174677 | standard |
| 2.74304 | 0.339614 | standard |
| 2.75647 | 0.174419 | standard |
| 3.82751 | 0.174339 | standard |
| 3.84098 | 0.338805 | standard |
| 3.85446 | 0.174338 | standard |
| 7.54208 | 0.286824 | standard |
| 7.54805 | 0.270525 | standard |
| 7.55589 | 0.260049 | standard |
| 7.57563 | 0.0119451 | standard |
| 7.9581 | 0.135061 | standard |
| 7.96229 | 0.14356 | standard |
| 7.97048 | 0.165481 | standard |
| 7.97633 | 0.110854 | standard |
| 7.97928 | 0.129614 | standard |
| 2.35567 | 1.0 | standard |
| 2.73184 | 0.174799 | standard |
| 2.74304 | 0.339938 | standard |
| 2.75424 | 0.174796 | standard |
| 3.82974 | 0.174786 | standard |
| 3.84093 | 0.340359 | standard |
| 3.85218 | 0.174285 | standard |
| 7.54331 | 0.295554 | standard |
| 7.54801 | 0.274848 | standard |
| 7.55471 | 0.249206 | standard |
| 7.5602 | 0.0866001 | standard |
| 7.57132 | 0.0127644 | standard |
| 7.9599 | 0.134433 | standard |
| 7.96324 | 0.147704 | standard |
| 7.97007 | 0.171154 | standard |
| 7.97752 | 0.130793 | standard |
| 2.35567 | 1.0 | standard |
| 2.73343 | 0.174662 | standard |
| 2.74304 | 0.339769 | standard |
| 2.75265 | 0.17466 | standard |
| 3.83132 | 0.174651 | standard |
| 3.84093 | 0.339887 | standard |
| 3.85054 | 0.174652 | standard |
| 7.54413 | 0.303649 | standard |
| 7.54791 | 0.281944 | standard |
| 7.5538 | 0.245089 | standard |
| 7.55909 | 0.0835315 | standard |
| 7.56824 | 0.0135031 | standard |
| 7.96107 | 0.132692 | standard |
| 7.96405 | 0.149682 | standard |
| 7.96985 | 0.172885 | standard |
| 7.97613 | 0.134228 | standard |
| 2.35567 | 1.0 | standard |
| 2.73412 | 0.174745 | standard |
| 2.74304 | 0.340063 | standard |
| 2.75206 | 0.174588 | standard |
| 3.83197 | 0.174886 | standard |
| 3.84093 | 0.340198 | standard |
| 3.84995 | 0.174577 | standard |
| 7.54448 | 0.30715 | standard |
| 7.54795 | 0.285494 | standard |
| 7.55344 | 0.24152 | standard |
| 7.55861 | 0.0827165 | standard |
| 7.56695 | 0.0138465 | standard |
| 7.96156 | 0.132215 | standard |
| 7.96435 | 0.152885 | standard |
| 7.96975 | 0.175197 | standard |
| 7.97563 | 0.134504 | standard |
| 2.35567 | 1.0 | standard |
| 2.73467 | 0.174822 | standard |
| 2.74309 | 0.340467 | standard |
| 2.75146 | 0.174638 | standard |
| 3.83251 | 0.17453 | standard |
| 3.84093 | 0.339382 | standard |
| 3.84936 | 0.174532 | standard |
| 7.54478 | 0.308414 | standard |
| 7.54792 | 0.289359 | standard |
| 7.55308 | 0.238361 | standard |
| 7.55809 | 0.0819941 | standard |
| 7.5659 | 0.0144292 | standard |
| 7.96207 | 0.13035 | standard |
| 7.9646 | 0.153137 | standard |
| 7.96967 | 0.174797 | standard |
| 7.97513 | 0.138007 | standard |
| 2.35567 | 1.0 | standard |
| 2.73561 | 0.17449 | standard |
| 2.74309 | 0.339675 | standard |
| 2.75057 | 0.174489 | standard |
| 3.83345 | 0.174832 | standard |
| 3.84093 | 0.339614 | standard |
| 3.84841 | 0.174833 | standard |
| 7.54528 | 0.305918 | standard |
| 7.5479 | 0.29847 | standard |
| 7.55255 | 0.234758 | standard |
| 7.55725 | 0.0796062 | standard |
| 7.5641 | 0.0153583 | standard |
| 7.96276 | 0.130383 | standard |
| 7.96499 | 0.15617 | standard |
| 7.96956 | 0.176521 | standard |
| 7.97445 | 0.139407 | standard |
| 2.35567 | 1.0 | standard |
| 2.73601 | 0.174474 | standard |
| 2.74309 | 0.340047 | standard |
| 2.75017 | 0.174447 | standard |
| 3.83385 | 0.174818 | standard |
| 3.84093 | 0.339263 | standard |
| 3.84802 | 0.174842 | standard |
| 7.54547 | 0.30321 | standard |
| 7.54793 | 0.298429 | standard |
| 7.55238 | 0.232171 | standard |
| 7.55693 | 0.0781148 | standard |
| 7.56341 | 0.015596 | standard |
| 7.96305 | 0.130984 | standard |
| 7.96515 | 0.157882 | standard |
| 7.96949 | 0.176053 | standard |
| 7.97416 | 0.13883 | standard |
| 2.35567 | 1.0 | standard |
| 2.73635 | 0.174757 | standard |
| 2.74309 | 0.339963 | standard |
| 2.74983 | 0.174756 | standard |
| 3.8342 | 0.174336 | standard |
| 3.84093 | 0.339182 | standard |
| 3.84767 | 0.174336 | standard |
| 7.54567 | 0.301577 | standard |
| 7.54794 | 0.303287 | standard |
| 7.55216 | 0.224943 | standard |
| 7.5566 | 0.077156 | standard |
| 7.56264 | 0.0162334 | standard |
| 7.96336 | 0.131052 | standard |
| 7.96531 | 0.159846 | standard |
| 7.96945 | 0.175987 | standard |
| 7.97387 | 0.139606 | standard |
| 2.35567 | 1.0 | standard |
| 2.73794 | 0.174916 | standard |
| 2.74309 | 0.340384 | standard |
| 2.74824 | 0.174913 | standard |
| 3.83573 | 0.174902 | standard |
| 3.84088 | 0.340361 | standard |
| 3.84609 | 0.174056 | standard |
| 7.54654 | 0.292147 | standard |
| 7.548 | 0.325381 | standard |
| 7.55126 | 0.204956 | standard |
| 7.55209 | 0.208218 | standard |
| 7.55547 | 0.0685324 | standard |
| 7.55969 | 0.0189589 | standard |
| 7.96453 | 0.132854 | standard |
| 7.96599 | 0.164442 | standard |
| 7.96943 | 0.156583 | standard |
| 7.9726 | 0.146479 | standard |