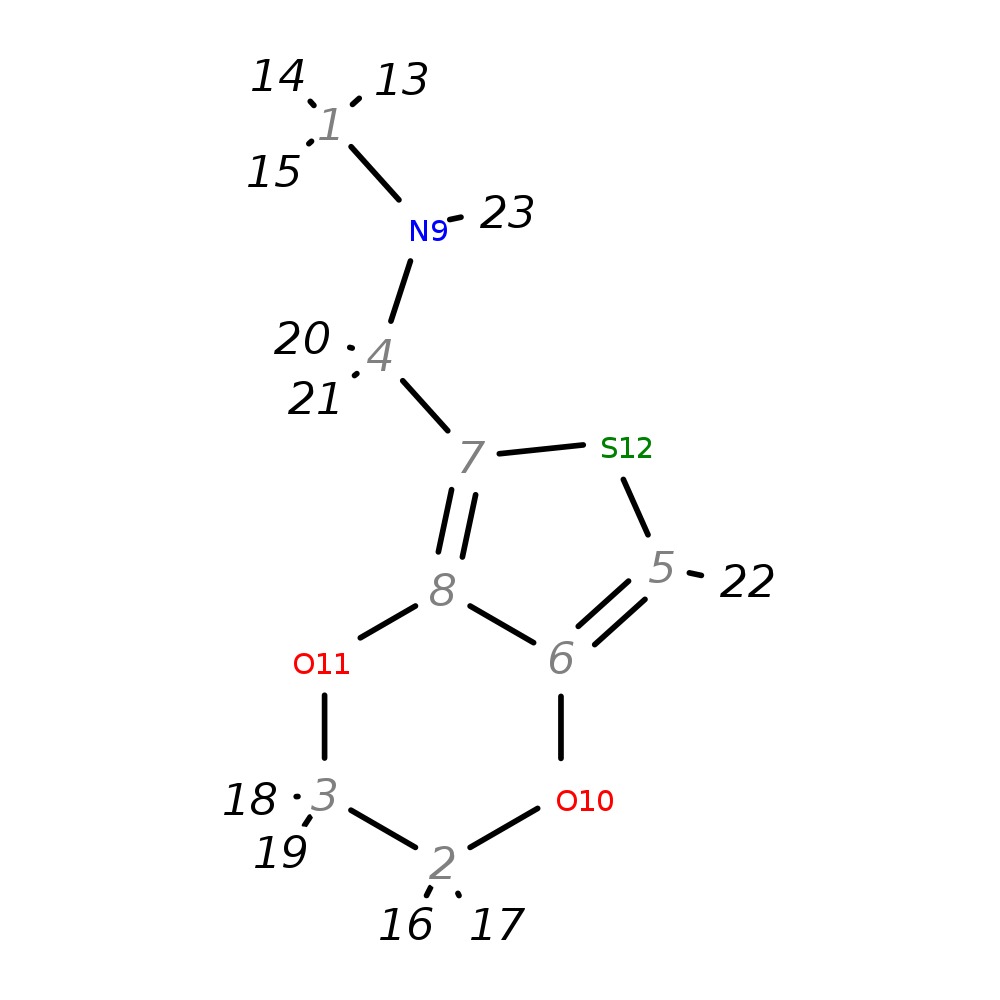
N-(2,3-dihydrothieno[3,4-b][1,4]dioxin-5-ylmethyl)-N-methylamine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01696 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H11NO2S/c1-9-4-7-8-6(5-12-7)10-2-3-11-8/h5,9H,2-4H2,1H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-(2,3-dihydrothieno[3,4-b][1,4]dioxin-5-ylmethyl)-N-methylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 2.697 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.697 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.697 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 4.305 | -14.0 | 5.837 | 5.758 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 4.305 | 6.619 | 6.537 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 4.267 | -14.0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 4.267 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.256 | -14.0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.256 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.617 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.69727 | 1.0 | standard |
| 4.2565 | 0.740925 | standard |
| 4.26681 | 0.255105 | standard |
| 4.2759 | 0.232774 | standard |
| 4.29636 | 0.22859 | standard |
| 4.30536 | 0.238607 | standard |
| 4.31505 | 0.0780369 | standard |
| 4.31777 | 0.0657881 | standard |
| 6.61751 | 0.333334 | standard |
| 2.69727 | 0.667622 | standard |
| 4.28092 | 1.0 | standard |
| 6.61751 | 0.222617 | standard |
| 2.69727 | 0.792388 | standard |
| 4.28435 | 1.0 | standard |
| 6.61751 | 0.263183 | standard |
| 2.69727 | 0.905766 | standard |
| 4.25907 | 0.861285 | standard |
| 4.28537 | 1.0 | standard |
| 6.61751 | 0.299483 | standard |
| 2.69727 | 0.960356 | standard |
| 4.25828 | 0.869091 | standard |
| 4.28559 | 1.00001 | standard |
| 6.61751 | 0.319791 | standard |
| 2.69727 | 1.0 | standard |
| 4.25779 | 0.870443 | standard |
| 4.28577 | 0.986585 | standard |
| 4.40967 | 0.0153391 | standard |
| 6.61751 | 0.333274 | standard |
| 2.69727 | 1.0 | standard |
| 4.22068 | 0.035153 | standard |
| 4.25662 | 0.734542 | standard |
| 4.26657 | 0.412148 | standard |
| 4.28409 | 0.555169 | standard |
| 4.28758 | 0.549946 | standard |
| 4.30435 | 0.30188 | standard |
| 4.33178 | 0.0520733 | standard |
| 4.35278 | 0.024646 | standard |
| 6.61751 | 0.330057 | standard |
| 2.69727 | 1.0 | standard |
| 4.23879 | 0.0551562 | standard |
| 4.25649 | 0.710013 | standard |
| 4.2667 | 0.319785 | standard |
| 4.2807 | 0.351146 | standard |
| 4.2913 | 0.348013 | standard |
| 4.30527 | 0.264046 | standard |
| 4.32316 | 0.077492 | standard |
| 4.33452 | 0.0351135 | standard |
| 6.61751 | 0.329191 | standard |
| 2.69727 | 1.0 | standard |
| 4.25651 | 0.738847 | standard |
| 4.26668 | 0.275322 | standard |
| 4.27905 | 0.271121 | standard |
| 4.29316 | 0.272466 | standard |
| 4.30537 | 0.246647 | standard |
| 4.31476 | 0.0755646 | standard |
| 4.31917 | 0.0702851 | standard |
| 4.32585 | 0.0458933 | standard |
| 6.61751 | 0.333299 | standard |
| 2.69727 | 1.0 | standard |
| 4.25644 | 0.733281 | standard |
| 4.26675 | 0.256024 | standard |
| 4.27709 | 0.250896 | standard |
| 4.29517 | 0.244767 | standard |
| 4.30537 | 0.239697 | standard |
| 4.31071 | 0.0954298 | standard |
| 4.31673 | 0.0726703 | standard |
| 4.32099 | 0.0556576 | standard |
| 6.61751 | 0.33329 | standard |
| 2.69727 | 1.0 | standard |
| 4.2565 | 0.739681 | standard |
| 4.26681 | 0.254686 | standard |
| 4.2759 | 0.232413 | standard |
| 4.29636 | 0.22826 | standard |
| 4.30536 | 0.238219 | standard |
| 4.31505 | 0.0779109 | standard |
| 4.31777 | 0.0656821 | standard |
| 6.61751 | 0.332055 | standard |
| 2.69727 | 1.0 | standard |
| 4.25648 | 0.741308 | standard |
| 4.26683 | 0.255839 | standard |
| 4.27476 | 0.226586 | standard |
| 4.2975 | 0.223194 | standard |
| 4.30536 | 0.24607 | standard |
| 4.31372 | 0.0837404 | standard |
| 4.31552 | 0.0747248 | standard |
| 6.61751 | 0.333333 | standard |
| 2.69727 | 1.0 | standard |
| 4.25652 | 0.726335 | standard |
| 4.26683 | 0.258138 | standard |
| 4.27437 | 0.215805 | standard |
| 4.29787 | 0.218306 | standard |
| 4.30536 | 0.249629 | standard |
| 4.31326 | 0.0867132 | standard |
| 4.31466 | 0.079267 | standard |
| 6.61751 | 0.333333 | standard |
| 2.69727 | 1.0 | standard |
| 4.2565 | 0.705215 | standard |
| 4.26683 | 0.261058 | standard |
| 4.27393 | 0.218008 | standard |
| 4.29833 | 0.215079 | standard |
| 4.30536 | 0.249871 | standard |
| 4.31278 | 0.0893786 | standard |
| 6.61751 | 0.333333 | standard |
| 2.69727 | 1.0 | standard |
| 4.25648 | 0.685205 | standard |
| 4.26678 | 0.269498 | standard |
| 4.27327 | 0.208069 | standard |
| 4.29896 | 0.21187 | standard |
| 4.30536 | 0.259949 | standard |
| 4.31205 | 0.0961842 | standard |
| 6.61751 | 0.333333 | standard |
| 2.69727 | 1.0 | standard |
| 4.25648 | 0.68052 | standard |
| 4.26678 | 0.275258 | standard |
| 4.27301 | 0.207526 | standard |
| 4.29925 | 0.205255 | standard |
| 4.30538 | 0.261467 | standard |
| 4.31181 | 0.100776 | standard |
| 6.61751 | 0.332333 | standard |
| 2.69727 | 1.0 | standard |
| 4.25648 | 0.67839 | standard |
| 4.26 | 0.145968 | standard |
| 4.26678 | 0.274867 | standard |
| 4.27275 | 0.208154 | standard |
| 4.29951 | 0.206101 | standard |
| 4.30536 | 0.265887 | standard |
| 4.31137 | 0.100624 | standard |
| 6.61751 | 0.333324 | standard |
| 2.69727 | 1.0 | standard |
| 4.25648 | 0.671763 | standard |
| 4.2621 | 0.129771 | standard |
| 4.26686 | 0.300167 | standard |
| 4.2715 | 0.203404 | standard |
| 4.30066 | 0.201578 | standard |
| 4.30534 | 0.285531 | standard |
| 4.3101 | 0.122638 | standard |
| 6.61751 | 0.33316 | standard |