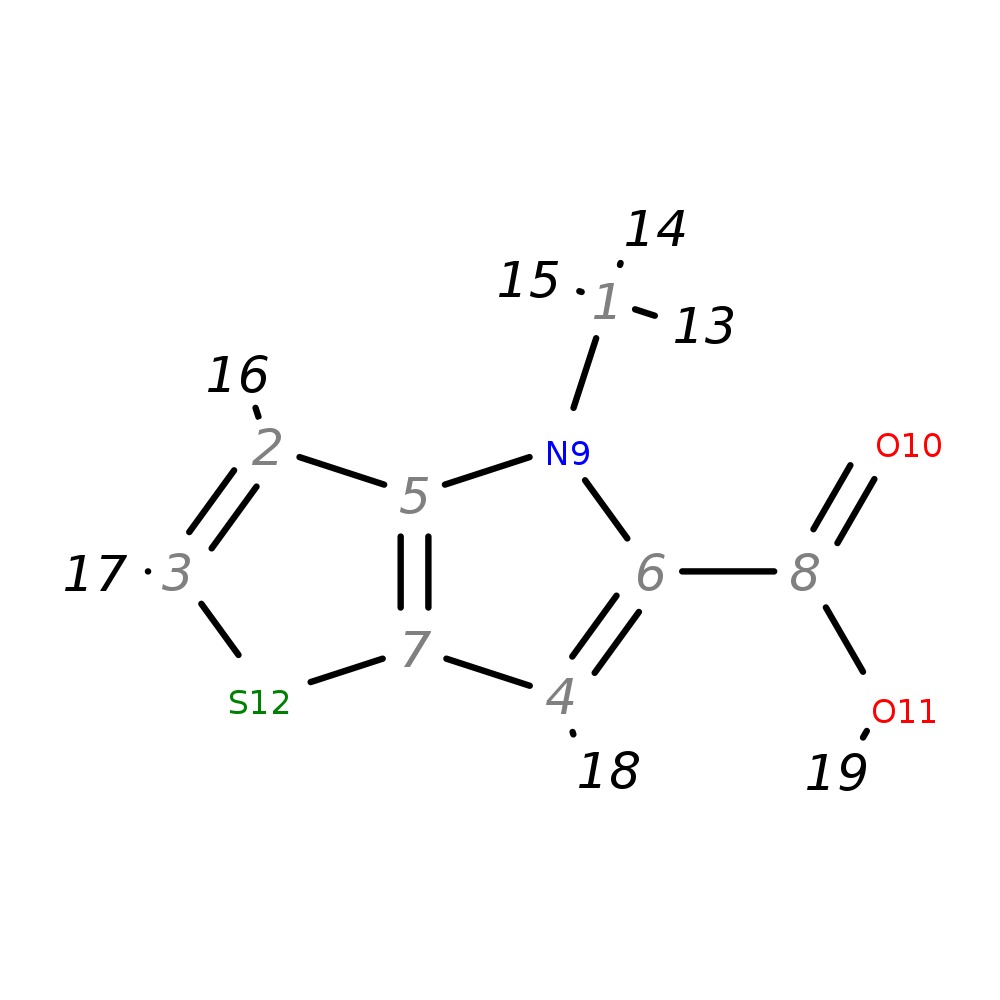
4-Methyl-4H-thieno[3,2-b]pyrrole-5-carboxylic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02353 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H7NO2S/c1-9-5-2-3-12-7(5)4-6(9)8(10)11/h2-4H,1H3,(H,10,11) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-Methyl-4H-thieno[3,2-b]pyrrole-5-carboxylic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|
| 13 | 3.972 | -14.0 | -14.0 | 0 | 0 | 0 |
| 14 | 0 | 3.972 | -14.0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 3.972 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 7.113 | 5.632 | 0 |
| 17 | 0 | 0 | 0 | 0 | 7.385 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 6.984 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333444 | standard |
| 7.10859 | 0.168208 | standard |
| 7.11789 | 0.175639 | standard |
| 7.38056 | 0.175593 | standard |
| 7.38986 | 0.168154 | standard |
| 3.97231 | 1.0 | standard |
| 6.98518 | 0.36363 | standard |
| 7.16655 | 0.258052 | standard |
| 7.3318 | 0.253795 | standard |
| 7.47228 | 0.100181 | standard |
| 3.97231 | 1.0 | standard |
| 6.98437 | 0.341142 | standard |
| 7.05815 | 0.135992 | standard |
| 7.15236 | 0.229271 | standard |
| 7.34606 | 0.227192 | standard |
| 7.43978 | 0.12016 | standard |
| 3.97231 | 1.0 | standard |
| 6.98431 | 0.33734 | standard |
| 7.07369 | 0.138231 | standard |
| 7.14394 | 0.215294 | standard |
| 7.3544 | 0.213631 | standard |
| 7.42467 | 0.132092 | standard |
| 3.97231 | 1.0 | standard |
| 6.98431 | 0.33595 | standard |
| 7.07857 | 0.140541 | standard |
| 7.14097 | 0.209891 | standard |
| 7.35747 | 0.209047 | standard |
| 7.41983 | 0.13587 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.335565 | standard |
| 7.08235 | 0.142788 | standard |
| 7.13859 | 0.206399 | standard |
| 7.35985 | 0.205356 | standard |
| 7.41607 | 0.139461 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333797 | standard |
| 7.09859 | 0.155819 | standard |
| 7.12661 | 0.188956 | standard |
| 7.37184 | 0.188621 | standard |
| 7.39986 | 0.155212 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333562 | standard |
| 7.10364 | 0.160937 | standard |
| 7.12235 | 0.18316 | standard |
| 7.3761 | 0.18299 | standard |
| 7.39481 | 0.160698 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333306 | standard |
| 7.10611 | 0.163488 | standard |
| 7.12017 | 0.18026 | standard |
| 7.37828 | 0.180161 | standard |
| 7.39234 | 0.16336 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333411 | standard |
| 7.1076 | 0.165777 | standard |
| 7.11878 | 0.178017 | standard |
| 7.37966 | 0.177964 | standard |
| 7.39085 | 0.165686 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333441 | standard |
| 7.10859 | 0.168207 | standard |
| 7.11789 | 0.175637 | standard |
| 7.38056 | 0.175591 | standard |
| 7.38986 | 0.168152 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333089 | standard |
| 7.10928 | 0.168653 | standard |
| 7.1173 | 0.174724 | standard |
| 7.38115 | 0.174989 | standard |
| 7.38917 | 0.168329 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333397 | standard |
| 7.10948 | 0.169924 | standard |
| 7.117 | 0.17364 | standard |
| 7.38145 | 0.173741 | standard |
| 7.38887 | 0.170023 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333389 | standard |
| 7.10977 | 0.170068 | standard |
| 7.1168 | 0.173552 | standard |
| 7.38164 | 0.173525 | standard |
| 7.38867 | 0.170038 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333395 | standard |
| 7.11017 | 0.170899 | standard |
| 7.11641 | 0.172756 | standard |
| 7.38204 | 0.172736 | standard |
| 7.38828 | 0.170875 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333366 | standard |
| 7.11037 | 0.171048 | standard |
| 7.11621 | 0.172805 | standard |
| 7.38224 | 0.172788 | standard |
| 7.38808 | 0.171026 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333358 | standard |
| 7.11047 | 0.170915 | standard |
| 7.11611 | 0.172592 | standard |
| 7.38234 | 0.172576 | standard |
| 7.38798 | 0.170895 | standard |
| 3.97231 | 1.0 | standard |
| 6.9843 | 0.333335 | standard |
| 7.11116 | 0.171925 | standard |
| 7.11542 | 0.171896 | standard |
| 7.38293 | 0.171696 | standard |
| 7.38729 | 0.171646 | standard |