Guided Ideographic Spin System Model Optimization (GISSMO)
GISSMO enables the efficient calculation and refinement of spin system matrices (chemical shift and coupling constants) against experimental 1D-1H NMR spectra of small molecules.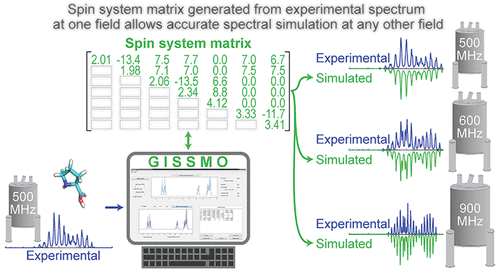 |
GISSMO library:
The library of parameterized compounds by GISSMO contains more than 1286 compounds, including many key mammalian
metabolites (list of biolocations) and a library of drug-like
molecular fragments used in ligand screening.
The library covers simulations of 1H NMR spectra at a variety of magnetic fields (40, 60, 80, 90, 100,
200, 300, 400, 500, 600, 700, 750, 800, 900, 950, 1000, 1100, and 1300 MHz) and in addition the library provides
spectral peak lists for every simulated spectra.
The exceptionally rich information content of NMR spectra is routinely used to identify and characterize molecules
in a wide range of applications, including clinical biomarker discovery, drug discovery, environmental chemistry,
and metabolomics. To facilitate seamless access to this reproducible and unique NMR metabolomics data library, every
entry in the GISSMO library can be downloaded in NMR-STAR
and NMReDATA data format.
The complete list of the entries in the GISSMO library can be accessed from
"Global links->GISSMO Library"
GISSMO GUI:
GISSMO utilizes a graphical user interface (GUI) for guided optimization of spin system matrices that can be downloaded from the “Global links->GISSMO-GUI”.
Citation:
- Hesam Dashti, Jonathan R. Wedell, William M. Westler, Marco Tonelli, David Aceti, Gaya K. Amarasinghe, John L. Markley, and Hamid R. Eghbalnia, Applications of Parametrized NMR Spin Systems of Small Molecules, Analytical Chemistry, 2018, DOI: 10.1021/acs.analchem.8b02660
- Hesam Dashti, William M. Westler, Marco Tonelli, Jonathan R. Wedell, John L. Markley, and Hamid R.
Eghbalnia, Spin System
Modeling of Nuclear Magnetic Resonance Spectra for Applications in Metabolomics and Small Molecule
Screening, Analytical Chemistry, 2017, 89 (22), pp 12201–12208, doi:
10.1021/acs.analchem.7b02884
Acknowledgement:
The GISSMO project is a collaboration between NMRFAM, NMRbox, and BMRB.
We are collaborating with the NMReDATA initiative and Mestrelab, and we are grateful for their
supports.
This study made use of the National Magnetic Resonance Facility at Madison, WI, which is supported by National
Institutes of Health (NIH) Grant P41GM103399. Metabolite data were from BMRB, which is supported by NIH Grant
GM109046. We are supported in part by the National Center for Biomolecular NMR Data Processing and Analysis,
which is supported by NIH Grant P41GM111135.